2582
Assessment of IDH1 Mutation Status and MGMT Promoter Methylation Status of Gliomas Using DWI, IVIM and DKI
Yan Xie1, Shihui Li1, Nanxi Shen1, Weiyin Vivian Liu2, and Wenzhen Zhu1
1Department of Radiology, Tongji Hospital, Tongji Medical College, HUST, Wuhan, China, 2MR Research, GE Healthcare, Beijing, China
1Department of Radiology, Tongji Hospital, Tongji Medical College, HUST, Wuhan, China, 2MR Research, GE Healthcare, Beijing, China
Synopsis
The purpose of this study was to evaluate the efficacy of conventional diffusion weighted imaging (DWI), intravoxel incoherent motion (IVIM) imaging and diffusion kurtosis imaging (DKI) in assessing IDH1 mutation status and MGMT promoter methylation status in gliomas. In lower-grade gliomas, diffusion MRI parameters were able to significantly distinguish the mutation status of IDH1 and the methylation status of MGMT promoter, which is helpful for predicting prognosis and sensitivity to alkylating chemotherapeutic agents of patients. However, in glioblastomas, there was no significant difference between mutant and wild-type IDH1 as well as methylation and unmethylation status of the MGMT promoter.
Introduction and Purpose
In 2016, the World Health Organization (WHO) classified levels of Central Nervous System (CNS) gliomas based on molecular features, specifically the isocitrate dehydrogenase 1 (IDH1) genotype1. It has been clinically demonstrated that alkylating agent showed good treatment efficacy on patients with the methylated O6-methylguanine-DNA methyltransferase (MGMT) promoter2. IDH1 mutation status and MGMT promoter methylation status are regarded clinically important indicators for tumor treatment and patient prognosis. However, the cost of gene screening is high and it is not easy to broadly implemented3. An imaging approach to provide not only anatomical details but also tissue function and expression is essential for clinic diagnosis. The purpose of our study was to assess IDH1 mutation degree and MGMT promoter methylation status of glioma using conventional diffusion weighted imaging (DWI), intravoxel incoherent motion (IVIM) imaging and diffusion kurtosis imaging (DKI).Materials and Methods
85 patients with pathologically confirmed gliomas underwent multi-b-value DWI with 20 b values (b= 0, 20, 50, 80, 100, 150, 200, 400, 600, 800, 1000, 1200, 1500, 2000, 2400, 2800, 3200, 3600, 4000 and 4500 s/mm2) and DKI (25 directions, b = 0, 1250, 2500 s/mm2) on 3.0T MRI (Discovery MR750, GE Healthcare). For DWI analysis, two b values (b=0, 1000) was used to compute conventional apparent diffusion coefficient (ADC). For IVIM analysis, two-segment mono-exponential and stretched-exponential models were used. One region of interest was placed on the most solid site of the tumor to obtain measurements including apparent diffusion coefficient (ADC), slow ADC (D), fast ADC (D*), perfusion fraction (f), distributed diffusion coefficient (DDC), heterogeneity index (α), fractional anisotropy (FA), mean diffusivity (MD) and mean kurtosis (MK). After the Shapiro-Wilk test and Levene test, the Independent Student’s t-test or Mann-Whitney U test was used to compare the differences of all parameters between IDH1 mutant and wild type as well as methylation and unmethylation status of the MGMT promoter in lower-grade gliomas and glioblastomas, respectively. Receiver operating characteristic (ROC) curves were performed to evaluate the discrimination performance of each parameter in distinguishing IDH1 genotype (mutant and wild-type) and MGMT status (methylation and unmethylation). The area under the receiver operating characteristic curve (AUC) was computed by DeLong test.Results
Among 49 patients with lower-grade glioma (grades II and III), 33 patients had mutant IDH1 genes and 16 patients had wild-type IDH1 genes, while the MGMT promoter was methylated in 22 patients and unmethylated in 27 patients. Among 36 patients with glioblastomas (grade IV), 6 patients had mutant IDH1 genes and 30 patients had wild-type IDH1 genes, whereas the MGMT promoter was methylated in 9 patients and unmethylated in 27 patients. In lower-grade gliomas, ADC, D, f, DDC, α and MD values were significantly higher in mutant IDH1 than in wild-type IDH1 (P <0.05) with AUC equal to 0.739, 0.750, 0.765, 0.748,0.780 and 0.759 (P <0.05), respectively. D* and MK values were significantly lower in mutant IDH1 than in wild-type IDH1 (P <0.05) with the AUC of 0.693 and 0.720 (p<0.05). ADC, D, f, DDC and MD values were significantly higher in MGMT promoter methylation than in unmethylated (P <0.05) with AUC equal to 0.704, 0.667, 0.678, 0.709 and 0.695 (P <0.05). Among glioblastomas, there was no significant difference of these parameters between mutant and wild-type IDH1 as well as unmethylated and methylated MGMT promoter.Discussion
Our study showed that ADC, D, D*, f, DDC, α, MD and MK had potential in accessing IDH1 mutation states in lower-grade gliomas. IDH1-mutant gliomas have a lower malignant biological behavior and a better prognosis than wild-type4. Therefore, we suspected that wild-type IDH1 existed in the lower-grade gliomas; when the degree of structural complexity in gliomas became higher, more abundant microvasculature and more diffusion barriers appeared. Mutant IDH1 in glioblastomas showed better prognosis than wild-type IDH1, which may be associated with a lower degree of cell proliferation5. ADC, D, f, DDC and MD values were significantly different between lower-grade gliomas with and without MGMT promoter methylation, but not in glioblastomas, which suggested that less abundant capillaries, lower cell density, larger inter-cell gaps and more restricted diffusion of water molecules were present in low-grade gliomas with methylated MGMT promoter. MGMT is a DNA repair protein found in the human body, and methylation of the MGMT promoter leads to inhibit expression of MGMT gene, reduce synthesis of MGMT protein, fail to repair damaged DNA and in the end apoptosis occurs6. Therefore, the diffusion and perfusion pattern in the highly malignant and complex structure in the glioblastomas was mixed and consequently it was difficult to reflect chaos with single parameter. This hnits us to further explore differences with the combination of multiple parameters in the future.Conclusion
In conclusion, diffusion MRI could predict the degree of IDH1 mutation and MGMT promoter methylation only in lower-grade gliomas, and thus it would be helpful in the prediction of prognosis and the planning of treatment. Moreover, IVIM and DKI do not show better performance on the prediction of IDH1 genotype and MGMT promoter methylation status compared to conventional DWI.Acknowledgements
Funding: This project was supported by the National Natural Science Funds of China (Grants No.81730049).References
- Karsy M, Guan J, Cohen AL, Jensen RL, Colman H. New Molecular Considerations for Glioma: IDH, ATRX, BRAF, TERT, H3 K27M. Curr Neurol Neurosci Rep. 2017;17(2):19.
- Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005; 352:997–1003.
- Jiang S, Rui Q, Wang Y, et al. Discriminating MGMT promoter methylation status in patients with glioblastoma employing amide proton transfer-weighted MRI metrics. Eur Radiol. 2018;28(5):2115-2123.
- Sabha N, Knobbe CB, Maganti M, et al. Analysis of IDH mutation, 1p/19q deletion, and PTEN loss delineates prognosis in clinical low-grade diffuse gliomas. Neuro Oncol. 2014;16(7):914-23.
- Yan W, Zhang W, You G, et al. Correlation of IDH1 mutation with clinicopathologic factors and prognosis in primary glioblastoma: a report of 118 patients from China. PLoS One. 2012;7(1):e30339.
- Jiang C, Kong Z, Liu S, et al. Fusion Radiomics Features from Conventional MRI Predict MGMT Promoter Methylation Status in Lower Grade Gliomas. Eur J Radiol. 2019;121:108714.
Figures
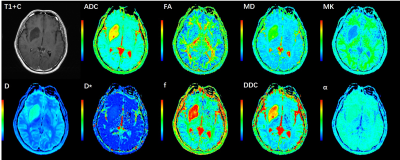
Figure 1 An illustration of a 39-year-old man with a lower grade glioma
(grade Ⅱ). IDH1 genotype was mutant and MGMT promoter status was methylated.
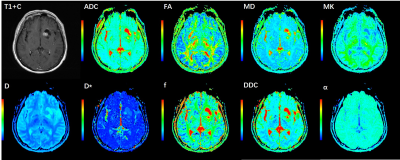
Figure 2 An illustration of a 57-year-old
man with a lower grade glioma (grade Ⅱ). IDH1 genotype was wild type and MGMT
promoter status was unmethylated.
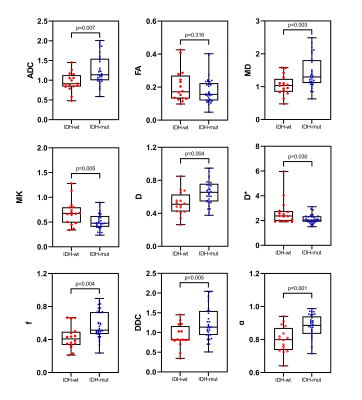
Figure 3 Box and whisker plots
of DWI, IVIM and DKI metrics in lower-grade
gliomas stratified according to IDH1 genotype (IDH1 mutant and IDH1 wild type).
Boxes represent the median ± quartiles, with whiskers extending to the maximum
and minimum values.
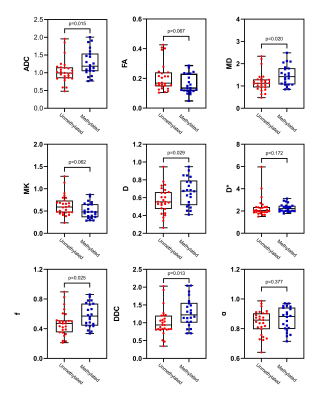
Figure 4 Box and whisker plots
of DWI, IVIM and DKI metrics in lower-grade
gliomas stratified according to MGMT promoter status (methylated and unmethylated).
Boxes represent the median ± quartiles, with whiskers extending to the maximum
and minimum values.
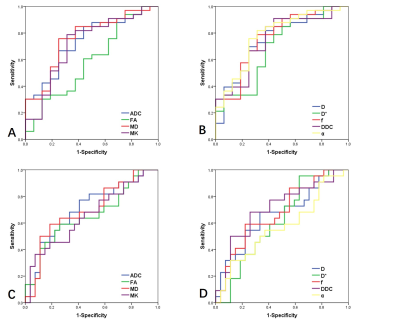
Figure 5 ROC curves for DWI, IVIM and DKI metrics to distinguish (A,
B) IDH1 genotype (IDH1 mutant and IDH1 wild type) as well as (C, D) MGMT promoter status
(methylated and unmethylated) in lower-grade gliomas.