2473
dMRIPrep: a robust preprocessing pipeline for diffusion MRI1The Centre for Addiction and Mental Health, Toronto, ON, Canada, 2Department of Psychology, University of Texas at Austin, Austin, TX, United States, 3eScience Institute, The University of Washington, Seattle, WA, United States, 4Basque Center on Cognition, Brain and Language, Donostia - San Sebastian, Spain, 5Neuroscience Program, University of Iowa, Iowa City, IA, United States, 6Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 7Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 8Department of Psychology, Stanford University, Stanford, CA, United States, 9NYU Grossman School of Medicine, New York City, NY, United States, 10Department of Psychology, The University of Washington, Seattle, WA, United States
Synopsis
We present dMRIPrep, a preprocessing pipeline for diffusion MRI (dMRI) inspired by the approach and wide uptake of fMRIPrep. dMRIPrep reliably and consistently performs on diverse data acquired by different studies. dMRIPrep equips researchers with a reliable and transparent tool developed with the best available engineering practices and neuroimaging standards, maintained as part of NiPreps, a community software framework that ensures long-lasting support and public-interest steering.
Introduction
The development and fast adoption of fMRIPrep1 have revealed that researchers need tools that simplify their workflow, provide visual reports with effective quality-control checkpoints, and are easily reproducible with proven reliability. dMRIPrep extends fMRIPrep's principles to diffusion MRI (dMRI). dMRI preprocessing requires many steps to clean and standardize the data before fitting a particular model or conducting tractography. Generally, researchers create ad-hoc preprocessing workflows for each dataset, building upon a growing inventory of available tools. Parallel to functional MRI2, the methodological variability across dMRI analysis workflows has snowballed with rapid advances in acquisition and processing. dMRIPrep is an analysis-agnostic tool that addresses the challenge of robust and reproducible preprocessing for whole-brain dMRI data.Methods
Implementation: dMRIPrep adopts the principles and software engineering best-practices of NiPreps3 (NeuroImaging PREProcessing toolS), a framework generalizing fMRIPrep's foundations to other neuroimaging modalities. The workflow only imposes one constraint on the input dataset - being compliant with BIDS4 (Brain Imaging Data Structure). BIDS-validity imposes formal consistency of data structuring and completeness of metadata. BIDS allows the workflow to minimize human intervention (no imaging parameters are manually entered) and to adapt itself to the unique properties of input data (e.g., making decisions about whether a particular preprocessing step is appropriate for this input data).dMRIPrep writes out BIDS-Derivatives, which extends BIDS specifications to the subproducts of processing and analysis algorithms (hence, derived data). By conforming to this standard, the derivatives of dMRIPrep are compatible with downstream methodologies (e.g., diffusion model fitting). Moreover, because the workflow does not execute preprocessing steps that bias results towards any particular analysis strategy, the pipeline is agnostic to further processing and does not limit downstream analysis choices.
Evaluation and validation: dMRIPrep is developed following fMRIPrep's acceptance and validation plan1. This includes two phases: Phase I is a fault discovery stage, including unit tests of individual preprocessing steps where a gold-standard is often available (e.g., a step with an analytic solution that can be independently tested) and integration tests of several of those steps. These integration tests are known as "smoke" tests, meaning that we only check whether the computational job exited with a success code and screen the reports for gross errors (e.g., a brain mask cutting out large areas of the brain). This phase includes feedback from the community through issue reports on our repository (https://github.com/nipreps/dmriprep). Phase II is a refinement stage, where simulated data and a larger sample of participants randomly selected from openly-available datasets (e.g., refs.5–8 and others) are benchmarked based on a range of signal-processing, compute-economy, and reproducibility metrics. Pipeline steps and their ordering are also compared to optimize performance in terms of accuracy and robustness.
Results
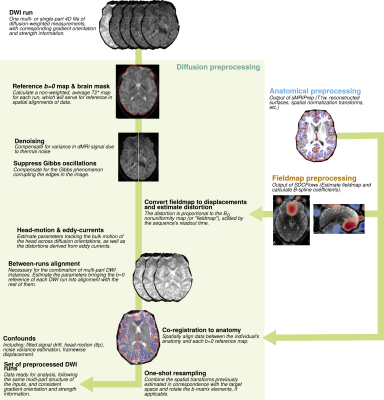
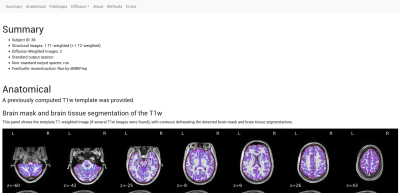
A reliable pipeline for the preprocessing of dMRI: Figure 1 illustrates the implementation of the pipeline. Each major step is defined as a Nipype workflow, which allows for modularity, creates well-defined inputs and outputs, and controls execution. For each set of dMRI scans, the diffusion gradient table is vetted and the orientation information of NIfTI headers is checked. A reference image and brain mask are calculated with b=0 volumes. Then, the estimation of several frequent imaging artifacts such as B1 inhomogeneity, noise modeling, and Gibbs oscillations is performed. Depending on the input dataset, available data to estimate B0 field maps are preprocessed with a shared component of NiPreps. Data deformation maps are derived from fieldmap information to account for susceptibility-derived distortions. Rigid-body head-motion parameters and eddy current-induced distortions are also estimated. In the case of multi-part dMRI sequences, data belonging to a single-run are aligned using the reference volumes calculated previously. The corresponding anatomical data is processed with a NiPrep and coregistered to the dMRI data. Finally, all spatial mappings (head-motion, eddy currents, susceptibility distortions, multi-part alignment, etc.) are concatenated and data are resampled in the designated output grid with a single interpolation step.dMRIPrep produces visual reports that allow quality-control of the results while serving as a scaffold for understanding the pipeline: Figure 2 shows an example of the visual report. A summary is visible at the top describing the subject ID, scans that were processed, and pipeline steps executed. The report exposes intermediate results through static and dynamic images. Finally, the report contains a citation boilerplate that describes each pipeline step and software version used. All of these elements seek to ensure the reproducibility of results and maximize transparency and self-documentation of the pipeline.
Discussion and conclusion
dMRIPrep equips dMRI researchers with a reliable and transparent tool developed with high standards of engineering and using cutting-edge neuroimaging methodologies, maintained with a community software model that ensures long-lasting support and public-interest steering. As with other NiPreps, dMRIPrep's goal is to provide "analysis-ready" data to other higher-level pipelines9–11 and serve as a data-workbench for researchers investigating new dMRI models and tractography, who can assume a level of consistency for the inputs of their algorithms. dMRIPrep democratizes dMRI analysis, as researchers can focus on modeling and analyzing their data, knowing that data preprocessing was performed in a robust and reproducible manner.Acknowledgements
No acknowledgement found.References
1. Esteban, O. et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116 (2019).
2. Botvinik-Nezer, R. et al. Variability in the analysis of a single neuroimaging dataset by many teams. Nature 582, 84–88 (2020).
3. Esteban, O. et al. NiPreps: enabling the division of labor in neuroimaging beyond fMRIPrep. in (2019). doi:10.31219/osf.io/ujxp6.
4. Gorgolewski, K. J. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044 (2016).
5. Magnotta, V. A. et al. MultiCenter Reliability of Diffusion Tensor Imaging. Brain Connect. 2, 345–355 (2012).
6. Hill, D., Williams, S., Hawkes, D. & Smith, S. M. IXI dataset - Information eXtraction from Images project (EPSRC GR/S21533/02). (Biomedical Image Analysis Group, Imperial College London, 2006).
7. Van Essen, D. C. et al. The Human Connectome Project: A data acquisition perspective. NeuroImage 62, 2222–2231 (2012).
8. O’Connor, D. et al. The Healthy Brain Network Serial Scanning Initiative: a resource for evaluating inter-individual differences and their reliabilities across scan conditions and sessions. GigaScience 6, (2017).
9. Yeatman, J. D., Richie-Halford, A., Smith, J. K., Keshavan, A. & Rokem, A. A browser-based tool for visualization and analysis of diffusion MRI data. Nat. Commun. 9, 940 (2018).
10. Cieslak, M. et al. QSIPrep: An integrative platform for preprocessing and reconstructing diffusion MRI. bioRxiv 2020.09.04.282269 (2020) doi:10.1101/2020.09.04.282269.
11. Tourbier, S. et al. Connectome Mapper 3: A Flexible and Open-Source Pipeline Software for Multimodal Human Connectome Mapping. Zenodo Softw. (2020) doi:10.5281/zenodo.3475969.