2448
Learning the relationship between human brain tissue microstructure and diffusion MRI data1Stanford, Stanford, CA, United States
Synopsis
We demonstrate the feasibility of machine learning direct prediction of tissue microstructure from raw diffusion MRI data. We test the approach by attempting to predict the main fiber orientation as this metric is well-understood and easily extracted using a diffusion tensor model. We present results on both simulated data and a matched dMRI-3D histology dataset.
Introduction
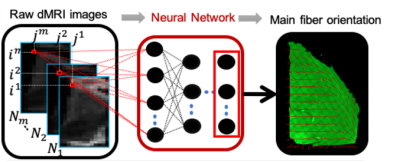
Diffusion MRI (dMRI) can be used to probe tissue microstructure non-invasively. This is usually achieved by performing a voxel-wise fit of the dMRI data to a biophysical model of diffusion and then extracting parameters that represent specific microstructural features such as axon diameter, density or dispersion. Unfortunately, the biophysical models employed often rely on strong assumptions and can be prone to overfitting and misinterpretation.[1] One approach for validating the resulting dMRI-derived microstructural parameter maps is through ex vivo experiments that directly compare MRI and histology results in the same specimen. [2-3] Here, we demonstrate the feasibility of a new approach to examine brain tissue microstructure with diffusion MRI: we applied deep learning techniques to learn the relationship between dMRI and microstructural features based on dMRI and 3D histology data in the same brain tissue (Figure 1). 3D histology offers a way to visualize microstructural features in 3D, by enabling histology of intact tissue cuboids[4-6]. We propose to leverage this type of data to learn the relationship between dMRI signals and tissue microstructure properties. To test the feasibility of the approach, we attempt to predict the main fiber orientation using 1) simulated dMRI and 2) high resolution dMRI and 3D histology data acquired on a localized deep gray matter region in the brain. The results obtained on the real dataset are then compared to the results obtained using the well-established and reliable DTI fit.Methods
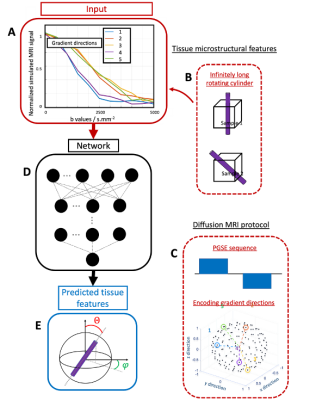
Simulated dataset: The diffusion MRI signal of a randomly orientated cylinder with a diameter of 4µm was simulated using the MATLAB MISST toolbox [7-10]. The chosen sequence was a Pulse Gradient Spin Echo (PGSE), 150 encoding gradient directions, 10ms duration and 5ms mixing time. We varied the b-value between 500 and 5000s/mm2 with a 500s/mm2 step size. The intrinsic diffusivity has been set to 0.4nm2/s which is consistent with postmortem tissues diffusivity[11]. The cylinder orientation was set using the polar angles theta and phi (Figure 2). The dMRI signal of 900 different cylinder orientations were simulated. Postmortem dMRI : MRI and CLARITY[4] were performed on a 1cm3 cuboid of human brain tissue containing part of the thalamus and part of the internal capsule (no known neuropathology, 4% PFA fixation). A diffusion MRI scan at 1mm isotropic resolution was acquired with b=2000s/mm2 and 450 diffusion directions. After MRI scanning, a 500μm thin sample was extracted from the tissue block and respectively cleared and stained with the CLARITY[4] and SWITCH protocols[12]. The sample cleared at 37°C for 2 weeks and was stained for nuclei and neurofilaments. The cleared sample was imaged with a fluorescence microscope to get an overview image and with a confocal microscope to get high-resolution images of the 3D volume. Extraction of the main fiber orientation from microscopy images: The dMRI/histology datasets were co-registered using a landmark-based registration in 3D Slicer[13]. The main fiber orientations of the histology images were extracted by measuring the structure tensors over regions corresponding to the MRI voxel resolution. Main fiber orientation prediction: A densely connected network using a root mean square loss function and Adam optimizer was trained to predict 1) the simulated cylinder orientation angles in polar coordinates and 2) the different components of the third eigenvector of the structural tensor of the histology images. The network was composed of 4 layers with ReLu activation function and a last linear layer. Dropout regularization was added after the first layer to avoid overfitting. The number of iterations was set to 3000 and 90% of the data (simulation: 810, real data: 81) were used to train the network (10 % of this subset was used for validation during training) whereas the 10% data left (simulation: 90, real data: 9) were used as the test set.Results and discussion
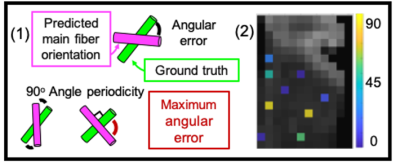
Using a simple densely connected network, the two cylinder spherical angles theta and phi could be predicted from the simulated signal with mean absolute errors of 15% and 5% which corresponds to errors of 13 and 6 degrees respectively. The simulations can be further extended to establish a framework to predict more complex microstructural features such as radius, dispersion or density from raw diffusion MRI data. For the dMRI and 3D histology data acquired in the same brain specimen, the mean angular difference between the fiber orientations derived from the MRI-based deep learning predictions and the 3D histology structure tensor estimates on the test set was 33+/-21 degrees (Figure 3). A similar angular difference was found between the fiber orientation estimates from MRI-based diffusion tensor estimates and 3D histology structure tensor estimates for the same 9 voxels was 35+/-16 degrees. These results inspire confidence that the dense network learned the features from the raw dMRI data that are necessary to predict the main fiber orientation.Conclusion
In this study, we demonstrated that it is possible to directly access tissue microstructure information from the raw diffusion MRI signal using both simulations and 3D histology images of intact human brain cuboids to train a deep learning network. Future efforts will compare the efficacy of more complex networks to learn additional features such as cell density or fiber dispersion in thicker tissue cuboids.Acknowledgements
The authors would like to thank the Wu Tsai Neuroscience Institute and the National Institute of Health (NIH) for their financial contribution to this project.References
[1] A. Rokem, J. D. Yeatman, F. Pestilli, K. N. Kay, A. Mezer, S. van der Walt, and B. A. Wandell. Evaluating the accuracy of diffusion mri models in white matter.PLOS ONE, 10(4):1–26, 04 2015.
[2] A. Seehaus, A. Roebroeck, M. Bastiani, L. Fonseca, H. Bratzke, N. Lori, A. Vilanova, R. Goebel, and R. Galuske. Histological validation of high-resolution dti in human post mortem tissue.Frontiers in Neuroanatomy, 9:98, 2015.
[3] K. G. Schilling, V. Janve, Y. Gao, I. Stepniewska, B. A. Landman, and A. W. Anderson. Histological validation of diffusion mri fiber orientation distributions and dispersion.NeuroImage, 165:200 – 221, 2018.
[4] K. Chung, J. Wallace, S.-Y. Kim, S. Kalyanasundaram, A. S. Andalman, T. J. Davidson, J. J. Mirzabekov, K. A.Zalocusky, J. Mattis, A. K. Denisin, H. Bernstein S. Pak, C. Ramakrishnan, L. Grosenick, V. Gradinaru, andK. Deisseroth. Structural and molecular interrogation of intact biological systems.Nature, 497:332337, 2013
[5] R. Renier, Z. Wu, D. J. Simon, J. Yang, P. Ariel, and M. Tessier-Lavigne. idisco: A simple, rapid method to immunolabellarge tissue samples for volume imaging.Cell, 159(4):896 – 910, 2014.
[6] Y.-G Park, C. H. Sohn, R. Chen, M. McCue, D. H. Yun, G. T. Drummond, T. Ku, N. B. Evans, H. C. Oak, W. Trieu, H. Choi,X. Jin, V. Lilascharoen, J. Wang, M. C. Truttmann, H. W. Qi, H. L. Ploegh, T. R. Golub, S.-C. Chen, M. P. Frosch, H. J. Kulik,B. K. Lim, and K. Chung. Protection of tissue physicochemical properties using polyfunctional crosslinkers.Nat. Biotech.,37:73–83, 2019.
[7] Ivana Drobnjak, Bernard Siow, Daniel C. Alexander, Optimizing gradient waveforms for microstructure sensitivity in diffusion-weighted MR, Journal of Magnetic Resonance,206(1): 41-51, 2010
[8] Ivana Drobnjak, Hui Zhang, Matt G. Hall, Daniel C. Alexander, The matrix formalism for generalised gradients with time-varying orientation in diffusion NMR, Journal of Magnetic Resonance, 210(1), 151-157, 2011
[9] Andrada Ianuş, Bernard Siow, Ivana Drobnjak, Hui Zhang, Daniel C. Alexander,Gaussian phase distribution approximations for oscillating gradient spin echo diffusion MRI,Journal of Magnetic Resonance, 27: 25-34, 2013
[10] Ianuş A., Alexander D.C., Drobnjak I. Microstructure Imaging Sequence Simulation Toolbox. In: Tsaftaris S., Gooya A., Frangi A., Prince J. (eds) Simulation and Synthesis in Medical Imaging. SASHIMI 2016. Lecture Notes in Computer Science, 9968, 2016
[11] Miller KL, Stagg CJ, Douaud G, et al. Diffusion imaging of whole, post-mortem human brains on a clinical MRI scanner. Neuroimage, 57(1):167-181, 2011.
[12] Evan Murray*, Jae Hun Cho*, Daniel Goodwin*, Taeyun Ku*, Justin Swaney*, Sung-Yon Kim, Heejin Choi, Jeong-Yoon Park, Austin Hubbert, Meg McCue, Young-Gyun Park, Sara Vassallo, Naveed Bakh, Matthew Frosch, Van J. Wedeen, H. Sebastian Seung, and Kwanghun Chung.Simple, scalable proteomic imaging for high-dimensional profiling of intact systems, Cell, Dec 3:163(6): 1500-14.
[13] Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J-C., Pujol S., Bauer C., Jennings D., Fennessy F.M., Sonka M., Buatti J., Aylward S.R., Miller J.V., Pieper S., Kikinis R. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magn Reson Imaging. 30(9):1323-41, 2012.
Figures