2439
Improved image quality with Deep learning based denoising of diffusion MRI data1GE Global Research, Niskayuna, NY, United States, 2GE Healthcare, New York, NY, United States, 3Walter Reed National Military Medical Center and Uniformed Services University of the Health Sciences, Bethesda, MD, United States, 4GE Healthcare, Menlo Park, CA, United States
Synopsis
Structure-preserved denoising of MRI images is a critical step in medical image analysis. This is particularly critical in diffusion MRI where higher spatial and angular resolutions required to map tissue microstructure in low SNR (especially at higher b-values) situations, if longer acquisition times are not used. Denoising using deep convolutional neural networks (DCNN) can reduce noise without requiring extensive averaging, enabling shorter scan times and high image quality, especially in the resulting tensor-derived maps. Preliminary results using DCNN based denoising on multi-shell diffusion data demonstrates improved image quality and reduced noise, without compromising on structural integrity and tensor derived metrics.
Introduction
Multi-shell diffusion MRI has informed our understanding of the brain, both at the level of local microstructure as well as distributed long-range network properties. Multiple b-values are used to study the angular properties of the diffusion signal, in particular the directionality of fiber bundles in white matter (WM). Typically, these high b-values result in lower signal-to-noise ratios (SNR), which can be compensated for by higher sampling (averaging) and longer scan times which are prohibitive in clinical scenarios. To compensate for low SNR, various signal denoising techniques have been proposed including low-rank approximations1, compressed sensing2 and model-based denoising methods3. Current unsupervised model-free denoising methods require a noise model to do the denoising, either as an explicit standard deviation and covariance, or implicitly in the choice of a noise correction method. Here we propose an unsupervised, model-free deep learning based denoising method that operates on the MR k-space raw data, eliminating the requirement to select or calibrate a noise model.Methods
Data from four acquisitions from two healthy volunteers were acquired under IRB-approval, on a whole-body 3T system (GE SIGNA MR750), equipped a high-performance head-gradient coil MAGNUS4,5, and using a 32-channel phased array head coil (NOVA Medical, Wilmington, MA). The MAGNUS gradient coil was operates at 200 mT/m with a maximum slew rate of 500 T/m/s for the diffusion gradients5. Multi-shell acquisitions were performed with either 300 (N=3 acquisitions) or 150 (N=1 acquisition) directions uniformly sampled over a sphere at b-values of 500, 1000, 2000, and 4000 s/mm2. Volunteer 1 was scanned twice with 300-direction diffusion acquisition and volunteer 2 was scanned twice with 300 and 150-direction diffusion acquisition. An additional 16 non-diffusion-weighted images (i.e., b=0) were interspersed through the acquisition. For a TR/TE = 44.2/7000 ms, 160x160 matrix, 86 slices, the 300 or 150-direction diffusion data were acquired with an isotropic resolution of 1.5x1.5x1.5 mm3. Diffusion images were denoised using commercially-available GE “AIRTM Recon DL” deep learning network6. Deep learning based denoising was performed at various intensities (50% and 75%) for comparison. The acquisition of the 300-direction data set allowed sub-sampling of the diffusion q-space to reduce the number of directions and provide the ability to characterize the denoising improvement compared to the longer scan with the large number of directions. All images (with or without DL based denoising) were post-processed using a custom -built pipeline. After distortion correction (rigid motion and eddy currents) using affine registration, standard diffusion tensor imaging parameters (fractional anisotropy (FA)) as well as diffusion kurtosis imaging parameters (parallel kurtosis (KPar), orthogonal kurtosis (KOrth)) were computed from the diffusion-weighted images. For WM parcellation, the individual subject images were non-rigidly registered to the FA atlas (John Hopkin’s ICBM DTI-81) with 50 parcellated WM regions, and the mean and standard deviation of all the diffusion maps within each of the parcels was computed.Results
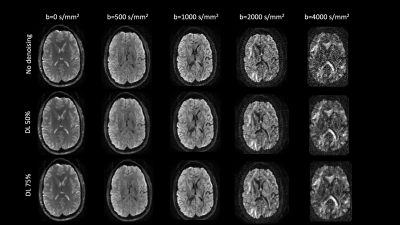
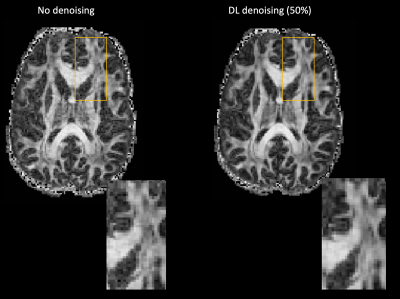
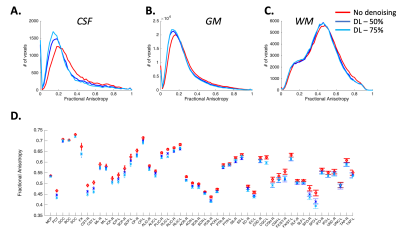
Qualitative assessment demonstrates superior performance of DL based denoising without compromising structural acuity especially at high b-values (Figure 1). Fractional anisotropy maps showed improved differentiation of signal components while reducing artifacts (Figure 2). Distribution of FA values across WM, Gray matter (GM) and CSF was computed (Figure 3A-C) to quantitatively assess the effect of DL-based denoising on the different signal compartments. While the distribution FA values in the whole-body WM was preserved, FA values in the DL-based denoised data were significantly lower than non-denoised images (Figure 3A) indicating a reduction of overall noise. Looking more closely, FA values in the individual WM parcels (50 WM parcels derived from the JHU-atlas) were preserved even after denoising, indicating that DL-based denoising did not alter or smooth quantitative diffusion maps. Tensor-derived diffusion metrics were compared between reference 300-direction and 150-direction diffusion data (volunteer 2) with and without DL denoising. While FA and KPar were not significantly different in the 150-direction compared to reference 300-direction data, KOrth values computed from DL denoised data were closer to reference values compared to non-denoised data.Conclusion and Discussion
Here we demonstrate initial results from a model-free DL-based denoising of diffusion weighted images and show reduced noise and improved Gray matter, White matter and CSF differentiation, without compromising quantitative diffusion metrics in individual WM regions. This DL-based approach does not require selection of a specific noise model and can potentially be used to accelerate acquisitions with minimal loss in quantitative diffusion metrics and robust structural connectivity.Acknowledgements
Grant funding from NIH U01EB28976, NIH U01EB024450, and CDMRP W81XWH-16-2-0054References
1. Veraart, J. et al. Denoising of diffusion MRI using random matrix theory. NeuroImage 142, 394–406 (2016).
2. Menzel, M. I. et al. Accelerated diffusion spectrum imaging in the human brain using compressed sensing. Magn. Reson. Med. 66, 1226–1233 (2011).
3. Sperl, J. I. et al. Model-based denoising in diffusion-weighted imaging using generalized spherical deconvolution. Magn. Reson. Med. 78, 2428–2438 (2017).
4. Foo, T. K. F. et al. Highly efficient head-only magnetic field insert gradient coil for achieving simultaneous high gradient amplitude and slew rate at 3.0T (MAGNUS) for brain microstructure imaging. Magn. Reson. Med. 83, 2356–2369 (2020).
5. Tan, E. T. et al. Peripheral nerve stimulation limits of a high amplitude and slew rate magnetic field gradient coil for neuroimaging. Magn. Reson. Med. 83, 352–366 (2020).
6. Lebel, R. M. Performance characterization of a novel deep learning-based MR image reconstruction pipeline. ArXiv200806559 Cs Eess (2020).
Figures