2436
A Comparative Study of Deep Learning Based Deformable Image Registration Techniques1Department of Biomedical Magnetic Resonance, Otto von Guericke University, Magdeburg, Germany, 2Data and Knowledge Engineering Group, Otto von Guericke University, Magdeburg, Germany, 3Faculty of Computer Science, Otto von Guericke University, Magdeburg, Germany, 4German Centre for Neurodegenerative Diseases, Magdeburg, Germany, 5Leibniz Institute for Neurobiology, Magdeburg, Germany, 6Center for Behavioral Brain Sciences, Magdeburg, Germany
Synopsis
Deep learning algorithms have been used extensively in tackling medical image registration issues. However, these methods have not thoroughly evaluated on datasets representing real clinic scenarios. Hence in this survey, three state-of-the-art methods were compared against the gold standards ANTs and FSL, for performing deformable image registrations on publicly available IXI dataset, which resembles clinical data. The comparisons were performed for intermodality and intramodality registration tasks; though in all the respective papers, only the intermodality registrations were exhibited. The experiments have shown that for intramodality tasks, all the methods performed reasonably well and for intermodality tasks the methods faced difficulties.
Introduction
The impressive performance of deep learning algorithms has inspired many to implement them in solving a multitude of real life scenarios like medical image registration1. In this survey, we have performed an analysis on three of the state-of-the-art methods2,3,4 for intermodality registration of the same subject but of different MR contrasts (T1 and T2) and intramodality registration of different subjects but of the same contrast (T1). In all these three papers, the experiments were limited to only intramodality registration. Here we tried to extend the analysis by performing intermodality experiments. All the experiments were performed in 3D. Final results were compared against the registrations performed by well-established tools, ANTs5 and FSL6.Methods
We have selected three state-of-the art algorithms: VoxelMorph3, ADMIR4, ICNet2. VoxelMorph takes a pair of affinely aligned moving and fixed images and calculates the deformation field by learning a set of parameters defined using a convolution neural network. ADMIR took this approach a step further and created an end-to-end pipeline where, first the raw images are transformed affinely, then it gets registered deformably. On the other hand, ICNet introduced two explicit constraints on the model namely inverse-consistent and anti-folding constraint, which helps to guide the model to generate more realistic deformations in comparison to those without these constraints, since DL methods are prone to generate unrealistic deformations and a strong regularizer is always required while defining the deformation function7.The experiments were performed on the publicly available IXI8 dataset. For the intermodel experiments T1 and T2 weighted images were used, whereas for intramodel experiments T1 weighted images were used. We used ANTs5,9 and FSL6,10 to pre-process our images before supplying them to the models. Pre-processing included skull-stripping, and affine registration2 3, both of which were done using FSL, and resampling which was done using ANTs. We tried to keep ourselves constrained to the implementation suggested by the authors, but in some places we have done some variations and/or experiments of our own, just to verify if we could get any better results by modifying or fine-tuning the models.The performance of the models were compared against the affine+deformable registrations performed with ANTs and FSL.
Experiments and Results
VoxelMorph3 model was trained on both raw and affinely registered, pre-processed images. The dataset consisted of 200 volumes and was trained for 1000 epochs. Here, we observed that VoxelMorph intramodal model couldn’t morph as expected if the fixed and moving images were not affinely aligned and pre-processed. There were some missing information and slight blurring in the warped images. On the other hand, VoxelMorph intramodal model on pre-processed images provided comparable results to that mentioned in the paper. We observed the brain folds of moving images deform as per fixed image and with more training, the blurring was reduced. VoxelMorph intermodal model was trained on pre-processed and affine registered images with similarity loss as mutual information. We observed that more bins led to better registration. Due to our limited computation power, we could only use 10 bins for mutual information loss and better registration results were seen here, albeit with slight blurring. Hence we believe that with more compute and bins, registration should work much better. Fig.1 shows the results of this method.As for ADMIR4, we were unable to stabilize the affine network, the initial stage of the pipeline, during training. The affine parameters caused too much shearing, and only the deformable part worked. As the coarsely warped image sheared too much, the correlation loss went to maximum causing too much load for the deformable network to correct it back. Furthermore, due to computational limitations, we could not perform ’intermodal’ registration for ADMIR, which is why there is no entry for it in the SSIM table (Tables.1). Results can be seen in Fig.2.
For ICNet2, as we increased the number of epochs and the dataset size for training, we noticed that the registration process was performing better. This only works for pre-affinely registered images. We can see from the figure that moving image A is trying to replicate fixed image B (result in A to B) for some parts of the brain. But, unfortunately, results are not as expected as we had hoped, mostly because complete warping did not take place and there was blurring in the result. But, if we consider the outcome of the ICNet paper, we realize that this result of ours can perform a lot better. Fig.3 shows the results obtained from this method.
The quantitative comparisons were performed with the help of SSIM11and has been exhibited in Table.1. For intramodal, it can be observed that VoxelMoroph is performing the best when compared against ANTs, and ADMIR when compared against FSL. For intermodal, The performance is very similar for both VoxelMorph and ICNet.
Conclusion
Our experiments showed that for the intramodal registrations, all the methods performed well. VoxelMorph achieved the highest SSIM (0.960) when compared against ANTs and ADMIR achieved the highest SSIM (0.959) when compared against FSL. For intermodal registrations, all the models had difficulties. Both VoxelMorph and ICNet performed similarly while being compared against ANTs (0.915 and 0.911 SSIM respectively), but ICNet performed better when compared against FSL (0.896).Acknowledgements
This work was in part conducted within the context of the International Graduate School MEMoRIAL at Otto von Guericke University (OVGU) Magdeburg, Germany, kindly supported by the European Structural and Investment Funds (ESF) under the programme "Sachsen-Anhalt WISSENSCHAFT Internationalisierung“ (project no. ZS/2016/08/80646).References
1. Grant Haskins, Uwe Kruger, and Pingkun Yan. Deep Learning in MedicalImage Registration: A Survey.Machine Vision and Applications, 31(1-2):8, February 2020. ISSN 0932-8092, 1432-1769. doi: 10.1007/s00138-020-01060-x. URLhttp://arxiv.org/abs/1903.02026. arXiv: 1903.02026.
2. Jun Zhang.Inverse-Consistent Deep Networks for Unsupervised De-formable Image Registration.arXiv:1809.03443 [cs], September 2018. URLhttp://arxiv.org/abs/1809.03443. arXiv: 1809.03443.
3. Guha Balakrishnan, Amy Zhao, Mert R. Sabuncu, John Guttag, and Adrian V. Dalca. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Transactions on Medical Imaging, 38(8):1788–1800, August 2019. ISSN 0278-0062, 1558-254X. doi:10.1109/TMI.2019.2897538.URLhttp://arxiv.org/abs/1809.05231.arXiv: 1809.05231.
4. Kun Tang, Zhi Li, Lili Tian, Lihui Wang, and Yuemin Zhu. ADMIR–Affine and Deformable Medical Image Registration for Drug-Addicted Brain Images. IEEE Access, 8:70960–70968, 2020. ISSN 2169-3536. doi: 10.1109/AC-CESS.2020.2986829.
5. Brian B Avants, Nick Tustison, and Gang Song. Advanced normalization tools (ants).Insight j, 2(365):1–35, 2009.
6. Stephen M Smith, Mark Jenkinson, Mark W Woolrich, Christian F Beckmann, Timothy EJ Behrens, Heidi Johansen-Berg, Peter R Bannister, Mar-ilena De Luca, Ivana Drobnjak, David E Flitney, et al. Advances in functional and structural mr image analysis and implementation as fsl. Neuroimage, 23:S208–S219, 2004.
7. Chen Qin, Shuo Wang, Chen Chen, Huaqi Qiu, Wenjia Bai, and Daniel Rueckert. Biomechanics-informed Neural Networks for Myocardial Mo-tion Tracking in MRI.arXiv:2006.04725 [cs, eess], July 2020.URLhttp://arxiv.org/abs/2006.04725. arXiv: 2006.04725.
8. IXIDatasetbybrain-development.org.URLhttps://brain-development.org/ixi-dataset/
9. AntsPy Plugin https://github.com/ANTsX/ANTsPy. Python implementation of ants tool. URLhttps://github.com/ANTsX/ANTsPy.
10. FSLhttps://fsl.fmrib.ox.ac.uk/fsl/fslwiki/.Fsltool.URLhttps://fsl.fmrib.ox.ac.uk/fsl/fslwiki/.
11. Z. Wang, A.C. Bovik, H.R. Sheikh, and E.P. Simoncelli. Image Quality Assessment: From Error Visibility to Structural Similar-ity. IEEE Transactions on Image Processing, 13(4):600–612, April2004. ISSN 1057-7149.doi:10.1109/TIP.2003.819861.URLhttp://ieeexplore.ieee.org/document/1284395/.
Figures
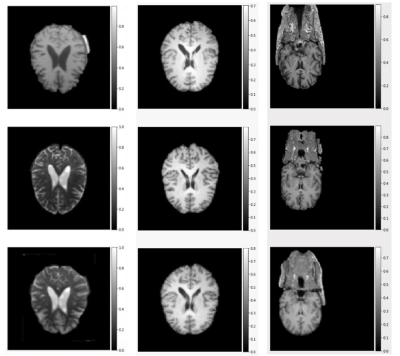
Fig.1: Result from experiments with VoxelMorph, after 1000 epochs.
Rows (top to bottom): fixed image, moving image, and warped image
Columns (left to right): Intermodal deformable registration of affinely registered pre-processed images, Intramodal deformable registration of affinely registered pre-processed images, and Intramodal registration performed directly on raw images without using any pre-processing.

Fig.2: Result from experiments with ADMIR, after 100 epochs.
Rows (top to bottom): fixed image, moving image, and warped image
Columns (left to right): Intramodal registration performed directly on raw images without using any pre-processing, and Intramodal deformable registration of affinely registered pre-processed images
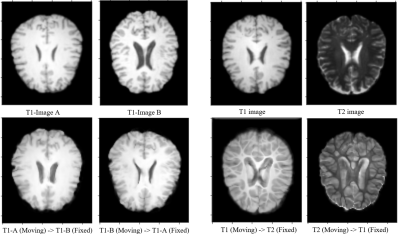
Fig.3: Result from experiments with ICNet.
(a) Intramodal deformable registration of affinely registered pre-processed images, after 100 epochs.
(b) Intermodal deformable registration of affinely registered pre-processed images, after 200 epochs.