2431
Multi-Contrast MRI Reconstruction from Single-Channel Uniformly Undersampled Data via Deep Learning1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong SAR, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong SAR, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
Synopsis
This study presents a deep learning based reconstruction for multi-contrast MR data with orthogonal undersampling directions across different contrasts. It enables exploiting the rich structural similarities from multiple contrasts as well as the incoherency arose from complementary sampling. The results show that the proposed method can achieve robust reconstruction for single-channel multi-contrast MR data at R=4.
Introduction
Routine clinical MRI sessions acquire multi-contrast images with identical geometries to maximize diagnostic information yet resulting in prolonged scan times. Images of different contrasts are often independently reconstructed despite their intrinsic anatomical similarities. The redundancy on the shared anatomical information can be utilized by jointly reconstructing multi-contrast images. Recent publications have demonstrated the benefit of jointly reconstructing images from MR data with multiple contrasts using deep learning (DL) to take advantage of the redundancy1.Conventional parallel imaging uniformly undersamples the k-space, this results in aliasing that manifests as coherent replicas of original image content. Additional calibration data is required to assist unfolding the aliasing. Incoherency across different contrasts can be introduced by orthogonally undersampling MR data of different contrasts, and exploited by jointly reconstructs multiple contrast MR data via low rank matrix completion2.
In this study, we propose a DL-based joint reconstruction method for single-channel multi-contrast MR data undersampled according to an orthogonally uniform pattern.
Methods
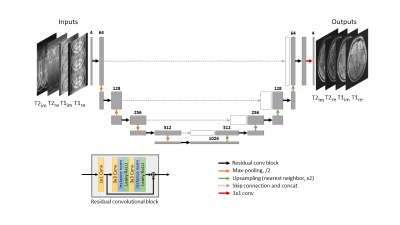
Model ArchitectureWe implemented a 2D Residual U-Net (Res-UNet) for jointly reconstructing the MR data with orthogonal undersampling directions across different contrasts. Res-UNet, as shown in Figure 1, is a U-Net framework with residual convolutional blocks. Real and imaginary components of complex T1- and T2-weighted, images were inputted as separate channels. The network was trained using Adam optimizer with initial learning rate 10-4, decay factor of 0.1 per 10 epochs, and l2 loss function. We trained the model for 30 epochs on a single GTX1080Ti.
Data Preparation
400 T1- and T2-weighted MR volumes from the HCP S1200 dataset3 were used for model training, validation, and testing. Multi-contrast MR data were prepared as follow.1) co-registering the T1- and T2-weighted MR volumes subject-by-subject using FSL FLIRT4, 2) downsampling the images by a factor of 2, resulting in identical in-plane geometry: FOV = 224×180 mm2 and resolution = 1.4×1.4 mm2, 3) adding different synthetic random 2D nonlinear phases to T1- and T2-weighted MR volumes separately, 4) applying orthogonal 1D uniform undersampling. The dataset was randomly split into training, validation and testing sets at a ratio of 8:1:1.
Evaluation
Experimental results were quantitatively evaluated using structural similarity index (SSIM) and normalized root mean square error (NRMSE). The performance of Res-UNet with complementary k-space sampling was evaluated with 1D acceleration at R = 3 and 4. The proposed method was also evaluated with images containing pathological regions.
Results
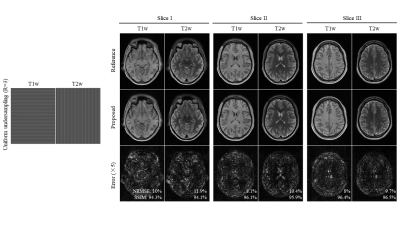
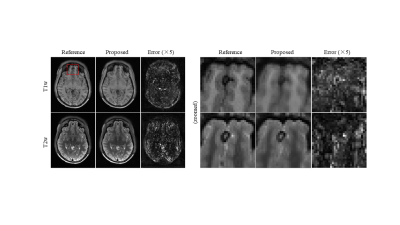
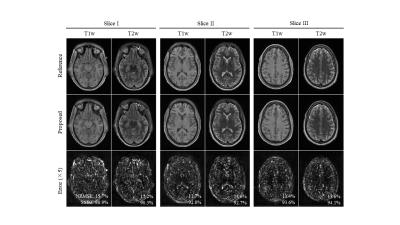
Figure 2 shows the reconstructed images for the proposed method at R=3. The proposed method successfully removes aliasing artefact introduced by the uniform undersampling. Figure 3 shows the robustness of the proposed multi-contrast MR reconstruction for brain images with pathology regions, where the correlations of different contrasts might differ from that in normal tissue. Figure 4 shows that the proposed method can achieve an undersampling factor of 4 with two contrasts jointly reconstructed using the proposed method.Discussion and Conclusion
In this study, we proposed a DL-based reconstruction for multi-contrast MR data, and demonstrated its effectiveness on single-channel MR dataset with T1w/T2w contrasts. The experimental results indicated that the proposed method could effectively remove the aliasing artifact at R=3. The results on pathological brain showed that our method, which was not specifically trained on pathology dataset, could reasonably reconstruct pathology. Orthogonally alternating PE direction increases the incoherency of aliasing caused by uniform undersampling. This incoherency allows similar reconstruction results to 2D random undersampling5. In this study, we demonstrated the joint reconstruction with orthogonal PE direction can also be realised via DL. Future studies will explore the use of patch based loss (e.g. SSIM6) instead of pixel-wise L2 loss, which may further improve the reconstruction in terms of reduced blurring effects.Acknowledgements
This study was supported by Hong Kong Research Grant Council (R7003-19, C7048-16G, HKU17112120, HKU17103819 and HKU17104020), Guangdong Key Technologies for Treatment of Brain Disorders (2018B030332001), and Guangdong Key Technologies for Alzheimer’s Disease Diagnosis and Treatment (2018B030336001).References
1. Polak D, Cauley S, Bilgic B, et al. Joint multi-contrast variational network reconstruction (jVN) with application to rapid 2D and 3D imaging. Magn Reson Med. 2020;84:1456–1469.
2. Yi Z, Liu Y, Zhao Y, Chen F, Wu EX. Joint calibrationless reconstruction of highly undersampled multi-contrast MR datasets using a novel low-rank completion approach. In27th Annual Meeting of ISMRM 2019. Montreal.
3. Van Essen, D.C., et al: The wu-minn human connectome project: an overview. Neuroimage 80, 62-79, 2013.
4. M. Jenkinson and S. Smith, “A global optimisation method for robust affine registration of brain images,” Med. Image Anal., 2001.
5. Wang H, Liang D, Ying L. Pseudo 2D random sampling for compressed sensing MRI. In: Conf Proc IEEE Eng Med Biol Soc, 2009, IEEE, 2672-2675.
6. Hammernik K, Knoll F, Sodickson DK, Pock T. L2 or not L2: Impact of Loss Function Design for Deep Learning MRI Reconstruction. In Proceedings of the International Society of Magnetic Resonance in Medicine (ISMRM). 2017. 0687
Figures