2428
Unsupervised deep learning for multi-modal MR image registration with topology-preserving dual consistency constraint1Paul C Lauterbur Research Center, Shenzhen Inst. of Advanced Technology, shenzhen, China, 2United Imaging Research Institute of Innovative Medical Equipment, Shenzhen, China
Synopsis
Multi-modal magnetic resonance (MR) image registration is essential in the clinic to achieve accurate imaging-based disease diagnosis and treatment planning. Although the existing registration methods have achieved good performance and attracted widespread attention, the image details may be lost after registration. In this study, we propose a multi-modal MR image registration with topology-preserving dual consistency constraint, which achieves the best registration performance with a Dice score of 0.813 in identifying stroke lesions.
Introduction
Multi-modal magnetic resonance (MR) imaging is one of the most prevalent techniques utilized in clinic as different MR imaging sequences can provide versatile information and highlight different regions of interest [1]-[3]. Comprehensive pathological information from multi-modal MR images provide a convenience for doctors to formulate more accurate and effective treatment plans. It is quite necessary to study the technology of multi-modal medical image registration.Deep learning has been a prevalent approach in medical image registration[4-8], making great progresses in multi-modal image analysis. Nevertheless, the performance of such methods can still be improved since few of them consider to preserve the topology of images.
This study design a deep learning-based topology-preserving schme, dual consistency constraint (DCC), to enhance the registration performance. A new inverse deformation field is calculated and applied to the forward registration images, we make similarity constraints between the obtained image and the original image, which can maximize the cross-correlation of topology maps of the registration images.
Method
With φ set as the pixels offset maps among different dimensions, each image’s deformation can be expressed as: MD = M ◦ φ, MD is the prediction of the moving image, M is the moving image, symbol ◦ refers to the transformation operator.The proposed dual consistency constraint refactor the φ firstly, as shown in Fig.1.For clarity, we decompose φ to obtain two offset fields φx and φy and then warp the offset fields with the original φ to form deformed offset fields. By recombining the deformed offset fields, a new transformation field is generated. Finally, the inverse transformation field φ-1 is obtained by multiplying with -1, symbol ◦ refers to the transformation operator, which consists of pixel shifting and interpolation.
$$\varphi^{-1}=-\sum_{x, y}\left(\varphi_{i} \circ \varphi\right)$$
∑ represents concatenating channels. φ-1 is directly derived from φ without any other operations.
Secondly, even if φ-1 is designed novelly, there still no accurate multi-modal registration loss function has been proposed yet, so the performance of the inverse transform may be limited. It is appropriate to transform the multi-modal registration into the single-modal, which means the inverse deformation is applied to the registration result instead of the fixed image, so as to obtain a topological advantage φ through reliable supervision. We use the prediction of moving image (MD) instead of the fixed image(F) to calculate the inverse transformed images: $$M^{\prime}=M_{D} \circ \varphi^{-1}$$ We assume M' should maintain the same distribution as M. Based on this, we use a consistency loss to accurate constraint M' to M, which can be MSE or NCC. We can obtain a more robust φ after updating the gradient in neural network, which can update the registration results. Besides, MI[9] loss is used to maximize the similarity between MD and F.
Dataset and Implementation
The proposed method has been evaluated on 555 patients (with or without stroke lesions) according to our ethics review board policy while each patient has given their consent. We used five sequences: T1 weighted, T2 weighted, FLAIR and DWI. All images were obtained with a Siemens 1.5T scanner. All the data are resized to 224×224 with intensity normalized [0, 1]. Our method is implemented using Keras with a Tensorflow backend on a NVIDIA Titan Xp GPU.Result and Discussion
Example registration result of different methods are shown in Fig.2. Though all methods can registerm in some way, there are big discrepancies. The red arrows indicate the regions where our method performs well but other methods failed to do so. Besides, we quantify the registration performance of stroke lesions by calculating three commonly used segmentation metrics (Dice score, precision, and recall). On one specific stroke area, as listed in table I. Compared to others, our method achieves the best Dice score of 0.813.In order to quantitatively analyze the effectiveness of the proposed dual consistency, we calculate the Jacobian determinant Jϕ (Table II). And |Jϕ| < 0 indicates the number of folded pixels, lower numbers indicate less deformation irregularities. By employing the proposed dual constraint, the mean number of locations with a non-positive jacobian determinants of each registration field is reduced from 227 to 74, proving that the constraint can effectively suppress the occurrence of folding deformations and preserve the topology of images.
In addition, our method is not limited to the registration of DWI and Flair modals, as shown in Fig.3. We register the MR images acquired with three functional sequences to DWI images using the proposed algorithm. Experiments prove that our method can work well.
Conclusion
Multi-modal MR imaging is critical for many clinical applications. Multi-modal misalignment often makes it difficult for doctors to accurately and quickly diagnose. Registration can alleviate these difficulties, but methods based on deep learning rarely consider the topology of the images. In this study, we propose a novel topology-preserving dual consistency strategy to achieve bi-directional image registrations. Compared to other registration methods, our method shows better registration performance in identifying stroke lesions.The proposed method is not limited to two specific modal images(Flair-DWI). More modal registrations can be mined, showing that the proposed method has good robustness. In the future, we will try more modal possibilities, including CT-MRI or PET-MRI. In general, our method shows good clinical application potential.
Acknowledgements
This research was partly supported by Scientific and Technical Innovation 2030-"New Generation Artificial Intelligence" Project (2020AAA0104100, 2020AAA0104105), the National Natural Science Foundation of China (61871371, 81830056), Key-Area Research and Development Program of GuangDong Province (2018B010109009), the Basic Research Program of Shenzhen (JCYJ20180507182400762), Youth Innovation Promotion Association Program of Chinese Academy of Sciences (2019351).
References
[1] G. Balakrishnan, A. Zhao, M. R. Sabuncu, J. Guttag, and A. V. Dalca, “An unsupervised learning model for deformable medical image registration,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 9252–9260.
[2] A. V. Dalca, G. Balakrishnan, J. Guttag, and M. R. Sabuncu, “Unsupervised learning for fast probabilistic diffeomorphic registration,” in International Conference on Medical Image Computing and ComputerAssisted Intervention. Springer, 2018, pp. 729–738.
[3] A. V. Dalca, G. Balakrishnan, J. Guttag, and M. R. Sabuncu, “Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces,” Medical image analysis, vol. 57, pp. 226–236, 2019.[4] M. Jaderberg, K. Simonyan, A. Zisserman, and k. kavukcuoglu, “Spatial transformer networks,” in Advances in Neural Information Processing Systems 28, C. Cortes, N. D. Lawrence, D. D. Lee, M. Sugiyama, and R. Garnett, Eds. Curran Associates, Inc., 2015, pp. 2017–2025. [Online]. Available: http://papers.nips.cc/paper/5854-spatial-transformer-networks.pdf
[5] G. Balakrishnan, A. Zhao, M. R. Sabuncu, J. Guttag, and A. V. Dalca, “An unsupervised learning model for deformable medical image registration,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 9252–9260.
[6] A. V. Dalca, G. Balakrishnan, J. Guttag, and M. R. Sabuncu, “Unsupervised learning for fast probabilistic diffeomorphic registration,” in International Conference on Medical Image Computing and ComputerAssisted Intervention. Springer, 2018, pp. 729–738.A. V. Dalca, G. Balakrishnan, J. Guttag, and M. R. Sabuncu, “Unsupervised learning for fast probabilistic diffeomorphic registration,” in International Conference on Medical Image Computing and Computer Assisted Intervention. Springer, 2018, pp. 729–738.
[7] G. Balakrishnan, A. Zhao, M. R. Sabuncu, J. Guttag, and A. V. Dalca, “Voxelmorph: a learning framework for deformable medical image registration,” IEEE transactions on medical imaging, vol. 38, no. 8, pp. 1788–1800, 2019.
[8] A. V. Dalca, G. Balakrishnan, J. Guttag, and M. R. Sabuncu, “Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces,” Medical image analysis, vol. 57, pp. 226–236, 2019.
[9] F. Maes, A. Collignon, D. Vandermeulen, G. Marchal, and P. Suetens, “Multimodality image registration by maximization of mutual information,” IEEE transactions on Medical Imaging, vol. 16, no. 2, pp. 187– 198, 1997.
Figures
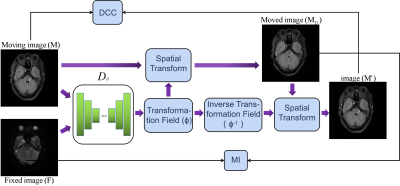
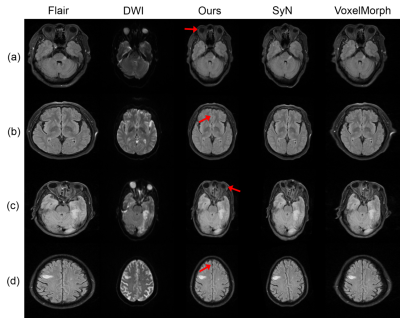
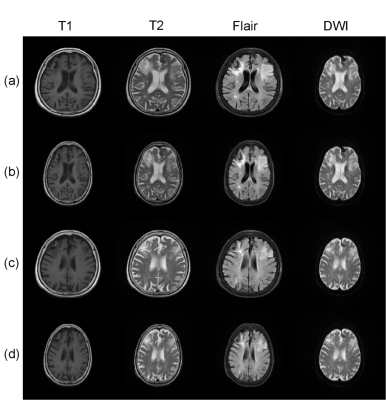
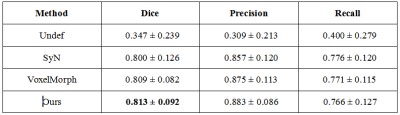
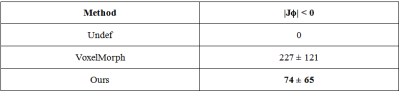
Table II Mean number of locations with a non-positive jacobian determinants of each registration field