2426
Multimodal Image Fusion Integrating Tensor Modeling and Deep Learning1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States
Synopsis
Multimodal brain imaging acquires complementary information of the brain. However, due to the high dimensionality of the data, it is challenging to capture the underlying joint spatial and cross-modal dependence required for statistical inference in various brain image processing tasks. In this work, we proposed a new multimodal image fusion method that synergistically integrates tensor modeling and deep learning. The tensor model was used to capture the joint spatial-intensity-modality dependence and deep learning was used to fuse spatial-intensity-modality information. Our method has been applied to multimodal brain image segmentation, producing significantly improved results.
Introduction
Multimodal brain imaging has become increasingly popular for both research and clinical applications.1 These multimodal images provide complementary information of brain tissues which, if used properly, can provide a more comprehensive understanding of brain function and disorders.2-4 However, due to the high dimensionality of multimodal image data, it is challenging to capture the underlying joint spatial and cross-modal dependence required for statistical inference in various brain image processing tasks. Conventional methods usually process each modality individually and then combine the results in the final stage,5 although efforts have been made to capture pairwise dependence between different modalities.6,7 Recently, deep learning-based methods have shown the potential to capture higher-order statistical dependence among multiple modalities.8,9 But those networks often require huge amounts of training data to avoid the over-fitting problem, which are often beyond what are currently available in various brain imaging applications. In this study, we proposed a new multimodal image fusion method, which synergistically integrates tensor modeling with deep learning. The tensor model was used to capture the joint spatial-intensity-modality dependence and deep learning was used to fuse intensity and spatial-intensity-modality information. While the proposed method is useful for a range of multimodal brain image processing tasks, including disease detection, classification and prediction, in this work, we focused on multimodal brain image segmentation to demonstrate its feasibility and potential.Method
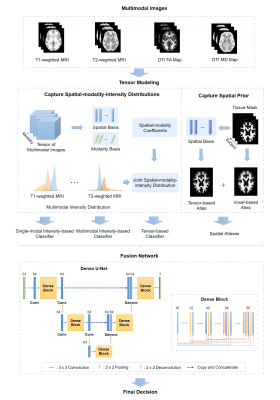
A fundamental problem in multimodal image fusion is to estimate the underlying joint probability distributions. This problem is challenging due to the difficulty associated with estimating high-dimensional probability functions and limited training data available. A common approach is to perform dimension reduction and/or assume some level of statistical independence of the multimodal data.10 In this work, a tensor model was used for dimension reduction and to capture the joint spatial-intensity-modality dependence.Tensor model as a dimension reduction transform
It is well known that a high-dimensional function, $$$I(x_1,x_2,…,x_d)$$$ can be well approximated by lower dimensional functions as 11:$$I(x_1,x_2,…,x_d)≈\sum_{l_1=1}^{L_1}\sum_{l_2=1}^{L_2}…\sum_{l_{\hat{d}}=1}^{L_{\hat{d}}}c_{l_1,l_2,…,l_{\hat{d}}}g_{l_1}^{{1}}({\bf{x}}_1)g_{l_2}^{{2}}{({\bf{x}}_2)}…g_{l_{\hat{d}}}^{{\hat{d}}}{({\bf{x}}_{\hat{d}})}\hspace{5cm}[1]$$where $$$\{{{\bf{x}}_1,{\bf{x}}_2,…{\bf{x}}_d ̂ }\}$$$ represents $$${\hat{d}}$$$ groups of “separable” variables from $$$\{{x_1,x_2,…,x_d}\}$$$. In our problem, we have $$$d={\hat{d}},\,x_1=x,\,x_2=y,\,x_3=z$$$, and $$$x_4=m$$$ that represents different modalities. Eq. [1] can be further expressed in a tensor form as:$$\mathcal{P}≈\mathcal{C}×_1G^{(1)}×_2G^{(2)}×…×_{\hat{d}}G^{({\hat{d}})}\hspace{5cm}[2]$$where $$$\mathcal{P}\in\mathbb{C}^{N_1×N_2×…×N_{\hat{d}}}$$$, $$$G^{(j)}∈\mathbb{C}^{N_j×L_j}$$$ and $$$\mathcal{C}\in\mathbb{C}^{L_1×L_2×…×L_{\hat{d}}}$$$ is the core tensor. We used the tensor model for two tasks: a) efficient representation of joint spatial-modality-intensity distributions; and b) dimension reduction of prior spatial probability distributions.
Task 1 includes three components: 1) determination of the tensor spatial-modality subspace; 2) estimation of the coefficients associated with the estimated subspace; and 3) joint intensity-spatial-modality distribution estimation. For the first problem, we applied high-order SVD to the training data to estimate the spatial-modality subspace as $$$\{{{\hat{G}}^{(i)}\}}_{i=1}^{{\hat{d}}-1}$$$. After that, the coefficients $$$w$$$ were obtained by projecting the multimodal data to the principal subspace such that $$${\hat{w}}={\hat{G}}^{({\hat{d}})}{\hat{C}}_{({\hat{d}})}={\hat{P}}_{({\hat{d}})}{\hat{Φ}}^{T}$$$, where $$${\hat{Φ}}=({\hat{G}}^{({\hat{d}}-1)}⊗{\hat{G}}^{({\hat{d}}-2)}⊗…⊗{\hat{G}}^{(1)} )^{T}$$$, $$${\hat{P}}_{({\hat{d}})}$$$ is the mode-$$${\hat{d}}$$$ matricization of $$$\mathcal{P}$$$, $$${\hat{C}}_{({\hat{d}})}$$$ is the mode-$$${\hat{d}}$$$ matricization of $$$\mathcal{C}$$$, and $$$⊗$$$ denotes the Kronecker product. Finally, the joint intensity-spatial-modality distribution was represented by the joint intensity and spatial-modality coefficients distribution using a mixture of Gaussians (MoG) model and the parameters could be estimated under the maximum likelihood principle.
For task 2, the prior spatial probability map of each brain tissue was generated from training data and was formed as a second-order tensor $$$\mathcal{T}\in\mathbb{C}^{N_1×N_2}$$$, which can be decomposed as: $$\begin{equation}\mathcal{T}≈\mathcal{S}×_1U^{(1)}×_2U^{(2)}\end{equation}\hspace{5cm}[3]$$ where $$$U^{(j)}∈\mathbb{C}^{N_j×L_j}$$$ represents the spatial/population basis of the model. The multimodal intensity distributions were projected to the estimated spatial-population subspace to get the tensor-based spatial statistical atlas The obtained joint spatial-modality-intensity distributions and spatial atlas were then integrated to build a tensor-based classifier.
Deep learning to fuse intensity and spatial-intensity-modality information
A Dense U-Net was used to integrate the following components: a) single-modal intensity-based classifiers; b) a multimodal intensity-based classifier; c) a tensor-based classifier; d) spatial atlases.12 The pipeline of our proposed fusion framework is illustrated in Figure 1.
Results and Discussion
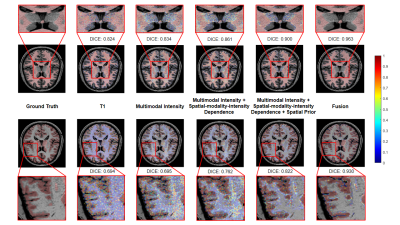
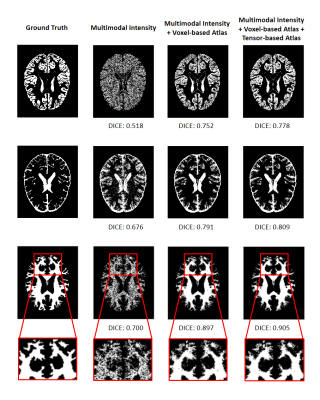
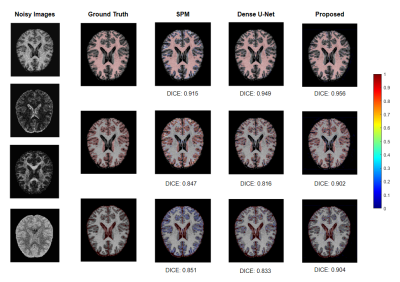
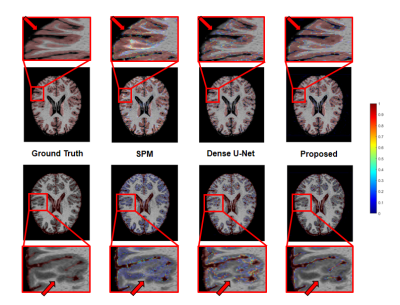
We applied our proposed fusion method to a multimodal brain tissue segmentation task and evaluate the performance using the Human Connectome Project (HCP) dataset.13 Gaussian noise was added to the original images to generate our testing data. Figure 2 shows a performance comparison by adding in each component in our fusion framework. The segmentation accuracy was improved with more features being added, which confirms the advantage of capturing spatial-intensity-modality dependence information. Figure 3 shows the advantage of combining the tensor-based spatial statistical atlas and the commonly used voxel-based spatial atlas. As can be seen, the segmentation performance was significantly improved after adding the tensor-based spatial atlas. Figure 4 compares the proposed method with other segmentation methods using SPM or Dense U-Net on noisy images.12,14 The proposed method produced more accurate segmentation results than the others. Figure 5 shows the tissue details of the segmentation results. As can be seen, our method performed much better in segmenting the cerebrospinal fluid branches.Conclusion
This paper presents a novel method for multimodal fusion, which synergistically integrates tensor modeling and deep learning. The proposed method was applied to multimodal brain image segmentation, producing significantly improved results over state-of-the-art methods. With further development, the proposed method can provide a powerful tool for many multimodal brain image processing tasks, including disease detection, classification and prediction.Acknowledgements
Y. L. is funded by National Science Foundation of China (No.61671292 and 81871083) and Shanghai Jiao Tong University Scientific and Technological Innovation Funds (2019QYA12).References
1. Menze BH, Van Leemput K, Lashkari D, et al. A generative probabilistic model and discriminative extensions for brain lesion segmentation—with application to tumor and stroke. IEEE Transactions on Medical Imaging. 2015;35(4):933-46.
2. Calhoun VD, Sui J. Multimodal fusion of brain imaging data: a key to finding the missing link (s) in complex mental illness. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. 2016;1(3):230-44.
3. Liu S, Cai W, Liu S, et al. Multimodal neuroimaging computing: a review of the applications in neuropsychiatric disorders. Brain Informatics. 2015;2(3):167.
4. Teipel S, Drzezga A, Grothe MJ, et al. Multimodal imaging in Alzheimer's disease: validity and usefulness for early detection. The Lancet Neurology. 2015;14(10):1037-53.
5. Vieira S, Pinaya WH, Garcia-Dias R, et al. Multimodal integration. Machine Learning. Academic Press, 2020;283-305.
6. Correa NM, Li YO, Adali T, et al. Canonical correlation analysis for feature-based fusion of biomedical imaging modalities and its application to detection of associative networks in schizophrenia. IEEE Journal of Selected Topics in Signal Processing. 2008;2(6):998-1007.
7. Mohammadi-Nejad AR, Hossein-Zadeh GA, Soltanian-Zadeh H. Structured and sparse canonical correlation analysis as a brain-wide multi-modal data fusion approach. IEEE Transactions on Medical Imaging. 2017;36(7):1438-48.
8. Plis SM, Amin MF, Chekroud A, et al. Reading the (functional) writing on the (structural) wall: multimodal fusion of brain structure and function via a deep neural network based translation approach reveals novel impairments in schizophrenia. Neuroimage. 2018;181:734-47.
9. Shi J, Zheng X, Li Y, et al. Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer's disease. IEEE Journal of Biomedical and Health Informatics. 2017;22(1):173-83.
10. Pan H, Liang ZP, Huang TS. Estimation of the joint probability of multisensory signals. Pattern Recognition Letters. 2001;22(13):1431-7.
11. He J, Liu Q, Christodoulou AG, Ma C, Lam F, Liang ZP. Accelerated high-dimensional MR imaging with sparse sampling using low-rank tensors. IEEE Transactions on Medical Imaging. 2016;35(9):2119-29.
12. Xiang L, Chen Y, Chang W, et al. Deep-learning-based multi-modal fusion for fast MR reconstruction. IEEE Transactions on Biomedical Engineering. 2018;66(7):2105-14.
13. Van Essen DC, Smith SM, Barch DM, et al. The WU-Minn human connectome project: an overview. Neuroimage. 2013;80:62–79.
14. Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26(3):839-51.
Figures