2424
Zero-dose FDG PET Brain Imaging1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States
Synopsis
PET is a widely used imaging technique but it requires exposing subjects to radiation and is not offered in the majority of medical centers in the world. Here, we proposed to synthesize FDG-PET images from multi-contrast MR images by a U-Net based network with symmetry-aware spatial-wise attention, channel-wise attention, split-input modules, and random dropout training strategy. The experiments on a brain tumor dataset of 70 patients demonstrated that the proposed method was able to generate high-quality PET from MR images without the need for radiotracer injection. We also demonstrate methods to handle potential missing or corrupted sequences.
Introduction
Positron emission tomography (PET) is a widely used molecular imaging technique with many clinical applications. To obtain high quality images, the amount of injected radiotracer in current protocols leads to the risks associated with radiation exposure in scanned subjects. Moreover, PET is expensive and not offered in the majority of medical centers in the world. MRI is a more widely available and non-invasive technique. Therefore, it is of great value to achieve zero-dose PET reconstruction, meaning synthesizing high-resolution and clinically-meaningful PET solely from multi-contrast MR images, which we achieve using convolutional neural networks with attention modules.Methods
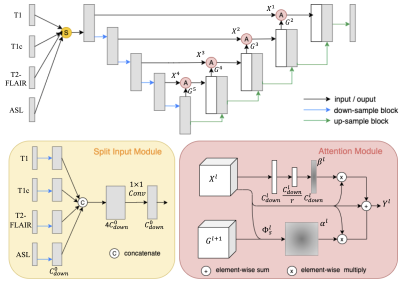
Data Acquisition and Preprocessing: With retrospective IRB approval, 70 patients with brain tumors who underwent clinically indicated FDG PET and MRI (T1, T1 with contrast (T1c), T2-FLAIR, and ASL) were included in this study. ASL perfusion imaging was first co-registered to PET. Then all images were co-registered to T1, placed in standard template space, and padded to size of 192 x 160 x 156. Intensities of the volumes was normalized by dividing by the mean of the non-zero regions. The top and bottom 20 slices were omitted. Three adjacent slices from each MR contrast were concatenated as the input. Random flipping of brain hemispheres was used as augmentation during training. Case-wise 5-fold cross-validation was conducted with 10% training cases used for validation.Model: As shown in Figure 1, a 2.5D U-Net worked as the backbone, on which spatial-wise and channel-wise attention modules were added to the short-cuts between layers. The symmetry-aware spatial-wise attention module (SSA) took features with a larger perceptive field from the lower upstream layer as the "guide" signal for learning attention for the detailed features from the downstream layer. In addition, given that the normal brain is roughly symmetric, radiologists tend to compare the left and right sides to take the advantage of asymmetry to help them identify abnormalities. We utilized the symmetry property by flipping the guided signal. The channel-wise attention module (CA) was adapted from the squeeze-and-excitation mechanism [1], factoring out the spatial dependency by global average pooling to learn a channel-specific value that can be used to re-weight the feature maps and emphasize the useful contrasts/channels. Moreover, as multi-contrast inputs may include redundant information and some contrasts are more or less important than others, we designed a split input module (SI) that first split each contrast, and then combined them with CA module before feeding into the U-Net. Lastly, we proposed a random dropout training strategy that simulated missing contrast scenarios to handle the incomplete multi-contrast inputs caused by data corruption or different imaging protocols in practice.
Results
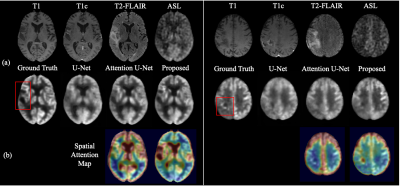
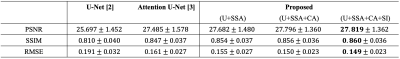
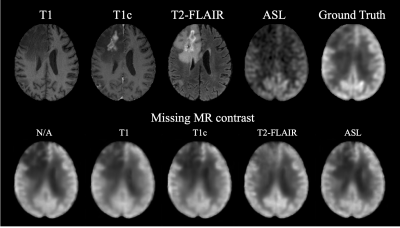
We compared our method with standard U-Net [2] and Attention U-Net [3] using the same backbone architecture and assessed by three metrics: peak signal-to-noise ratio (PSNR), structural similarity (SSIM), and root mean square error (RMSE). As shown in Table 1, the proposed method achieved highest image quality of PSNR 27.8, SSIM 0.860, and RMSE 0.149, improving 8.26% in PSNR, 6.17% in SSIM, and 21.99% in RMSE from U-Net. Moreover, we conducted an ablation study by gradually adding each proposed component to the backbone. The improving quality metrics demonstrated the effectiveness of the attenuation and split input modules. Qualitatively, two typical examples are shown in Figure 2. All three methods performed similarly on the normal regions of the brain. However, the proposed method synthesized images with more accurate pathological features on abnormal regions. Spatial attention maps from the second layer in Figure 2(b) illustrate that the proposed method precisely highlighted the tumor region, while maps from the Attention U-Net had weak signals, which led the poorer synthesis. Moreover, a radiologist was asked to diagnose the recurrence of tumor from the ground truth and synthesized PET for 10 cases that were equally and randomly selected from each fold. The congruent reading results illustrate that the proposed method could achieve high-quality and clinically-meaningful PET. Figure 3 demonstrates the effects of the random input dropout strategy. Though not as good as model without input dropout, it achieved synthesized images with slight changes except for the scenario that is missing T2-FLAIR, which might indicate that the model relies heavily on T2-FLAIR.Discussion
Compared with the widely used standard and simple attention U-Nets, the proposed method can generate high quality PET images with more precise details, including abnormalities. The image quality metrics of the ablation study illustrates the contribution of each proposed component. The spatial attention maps address the interpretability of the attention module. The reading result shows that the proposed method could synthesize high-quality and clinically-meaningful PET. The qualitative results with missing contrasts demonstrate that the proposed input dropout strategy achieves robust synthesis.Conclusion
We propose a U-Net based network with symmetry-aware spatial attention module to capture the abnormal region of the brain, and split-input with channel-wise attention module to enhance the most important features, as well as a random dropout training strategy to handle missing input contrasts. The results on the brain tumor dataset illustrate the potential of synthesize high-quality FDG-PET images from multi-contrast MR images without the use of radiotracers.Acknowledgements
Funding for this study was received form U.S. National Institutes Health (NIH) grants R01-EB025220, P41-EB015891. This study also benefited from GE Healthcare.References
1. Hu, Jie, Li Shen, and Gang Sun. "Squeeze-and-excitation networks." Proceedings of the IEEE conference on computer vision and pattern recognition. 2018.
2. Oktay, Ozan, et al. "Attention u-net: Learning where to look for the pancreas." Proceedings of the IEEE Medical Imaging with Deep Learning. 2018.
3. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
Figures