2418
Federated Multi-task Image Classification on Heterogeneous Medical data with Privacy Perversing1Monash Biomedical Imaging, Monash University, Australia, Melbourne, Australia, 2Monash eResearch Center, Monash University, Australia, Melbourne, Australia
Synopsis
There is a lack of pre-trained deep learning model weights on large scale medical image dataset, due to privacy concerns. Federated learning enables training deep networks while preserving privacy. This work explored co-training multi-task models on multiple heterogeneous datasets, and validated the usage of federated learning could serve the purpose of pre-trained weights for downstream tasks.
Introduction
Using pre-trained models from large public datasets has been a standard paradigm to tackle conventional computer vision tasks 1,2. The state-of-the-art performance of downstream tasks, e.g. image classification and segmentation are achieved by fine-tuning models from pre-trained weights trained on datasets like ImageNet and CoCo 3,4. However, in the field of medical image analysis, applying the model weights pre-trained with natural images failed to improve the model quality 5,6. Also, annotated medical imaging datasets always contain sensitive and privacy information, which makes it challenging to aggregate large scale public datasets, like ImageNet, and have an universal feature encoder that can be used in the downstream medical image analysis tasks.Federated Learning(FL) is a paradigm and technology stack that enables training shared machine learning models using datasets reside on multiple nodes (e.g. organizations) while preserving privacy 7. The technology has been used in cross-device settings, e.g. language modeling 8-10, and cross-silo settings, e.g. image analysis 11,. Some works discussed FL applications in medical imaging, however, they mainly focused on modeling single tasks 12-14. In this work, we applied federated multi-task learning, and showed the feasibility to pre-train large scale medical image models without the need to explicitly aggregate and access the private datasets.
Methods
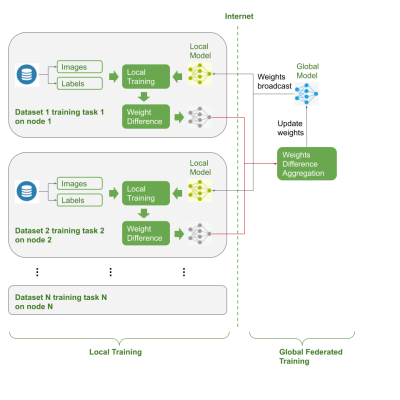
The dataset used in the study is the “Medical MNIST'', which contains 10 different image modalities (e.g. retina images, breast CT images etc.) and predefined tasks (inc. binary classification, multi-label classification and regression) 15. Each sub-dataset has a different number of image samples (from scale of 100 to 100,000) and classification categories, which are splitted into training, validation and test datasets.The federated environment was simulated to mimic the real-life FL implementations, where 10 federated nodes / clients were represented by the corresponding neural network models and private datasets. The training, validation and testing data were only accessible to its corresponding federated neural network and optimizer. The network was composed of a ResNet18 as the backbone and multiple linear layers as the classifier. As shown in Figure 1, the training process included: a) an initialized global model was broadcasted to all the federated nodes; b) each local model copied the shared global weights and initialized the local weights of the ResNet18, while neck layers, i.e. the classifier linear layers were independent from the global model; c) each federated nodes performed local optimization iterations; d) after each federated round (a predefined hyper parameter), the model updates were calculated by computing the difference between the latest weights and the weights synchronized from the global model at the beginning of the federated round; e) the weights updates from all the federated nodes were aggregated and averaged; f) the averaged weights updates were applied to the global model to complete one federated round. The process repeated to reach model convergence.
To validate the framework, accuracy (ACC) and Area Under Curve (AUC) were measured under different experimental conditions: i) each dataset was trained separately and measured as baseline metrics; ii) models were trained under FL setups, with different number of local iterations, including 10, 20 and 30; iii) performance were measured with per-sample differential privacy, to exam the privacy-performance tradeoffs.
Results and Discussion
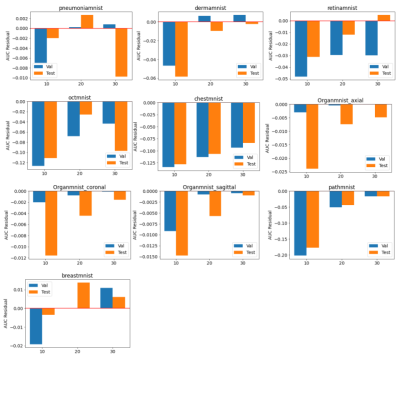
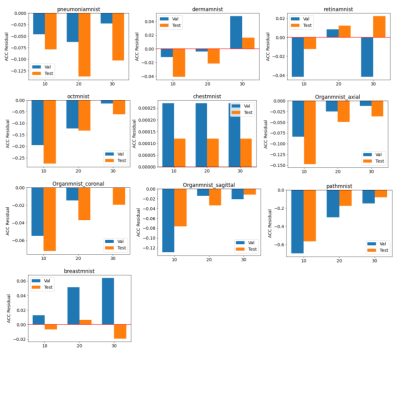
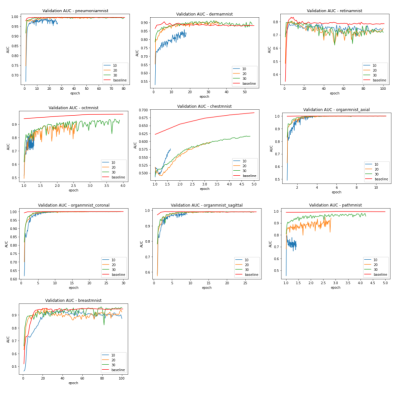
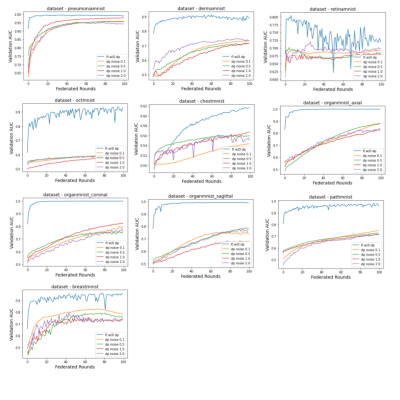
The baseline models were trained for 100 epochs on each dataset and the performance was measured separately. The results were very close to the numbers reported by the original dataset paper (ref). The standard parameters were used in the training, e.g. batch size of 128, standard SGD optimizer and learning rate of 0.001. The federated training applied the similar hyperparameter, while varied the number of local iterations in each federated round.As shown in figure 2 and 3, compared to the baseline models, federated trained multi-task models were performed slightly worse but similar in most of the datasets, e.g. the Pneumonia, Organ, Breast and Retina datasets, while a slight better performance (>1% in AUC) could be observed in the small Breast dataset, and the Retina dataset (>1% ACC). This indicated that the model trained with a small dataset obtained most of the benefits when involved in the federated training, even though the gradient signals were significantly weakened in the federated averaging process. Also, using a larger number of local iterations achieved better results, which was consistent among most of the datasets. In figure 4, it showed that more local iterations also lead to faster convergence.
To further improve the data privacy, a per-sample differential privacy (DP) mechanism was applied to each local iteration 16. However, Figure 5 showed that it might not be a good option in the multi-task learning setup, as the performance was dramatically worse. This suggested that per-sample DP might be the way to achieve data privacy in a single node setup, but not a federated learning training environment. In the later situation, adding DP to each federated round might be sufficient to preserve privacy, and achieve reasonable privacy versus utility tradeoffs.
Conclusion
In this work, we introduced and validated an noval federated multi-task learning framework that enabled co-training an universal feature encoder without explicitly accessing the datasets. It opens the possibility to transfer learning from federated trained models on large scale medical images that are privately owned by multiple entities.Acknowledgements
No acknowledgement found.References
1. Huh, M., Agrawal, P., & Efros, A. A. (2016). What makes ImageNet good for transfer learning?. arXiv preprint arXiv:1608.08614.
2. Studer, L., Alberti, M., Pondenkandath, V., Goktepe, P., Kolonko, T., Fischer, A., ... & Ingold, R. (2019, September). A comprehensive study of ImageNet pre-training for historical document image analysis. In 2019 International Conference on Document Analysis and Recognition (ICDAR) (pp. 720-725). IEEE.
3. Deng, J., Dong, W., Socher, R., Li, L. J., Li, K., & Fei-Fei, L. (2009, June). Imagenet: A large-scale hierarchical image database. In 2009 IEEE conference on computer vision and pattern recognition (pp. 248-255). Ieee.
4. Lin, T. Y., Maire, M., Belongie, S., Hays, J., Perona, P., Ramanan, D., ... & Zitnick, C. L. (2014, September). Microsoft coco: Common objects in context. In European conference on computer vision (pp. 740-755). Springer, Cham.
5. He, K., Girshick, R., & Dollár, P. (2019). Rethinking imagenet pre-training. In Proceedings of the IEEE international conference on computer vision (pp. 4918-4927).
6. Zoph, B., Ghiasi, G., Lin, T. Y., Cui, Y., Liu, H., Cubuk, E. D., & Le, Q. (2020). Rethinking pre-training and self-training. Advances in Neural Information Processing Systems, 33.
7. Kairouz, P., McMahan, H. B., Avent, B., Bellet, A., Bennis, M., Bhagoji, A. N., ... & d'Oliveira, R. G. (2019). Advances and open problems in federated learning. arXiv preprint arXiv:1912.04977.
8. Ramaswamy, S., Mathews, R., Rao, K., & Beaufays, F. (2019). Federated learning for emoji prediction in a mobile keyboard. arXiv preprint arXiv:1906.04329.
9. Yang, T., Andrew, G., Eichner, H., Sun, H., Li, W., Kong, N., ... & Beaufays, F. (2018). Applied federated learning: Improving google keyboard query suggestions. arXiv preprint arXiv:1812.02903.
10. Chen, M., Mathews, R., Ouyang, T., & Beaufays, F. (2019). Federated learning of out-of-vocabulary words. arXiv preprint arXiv:1903.10635.
11. Zhang, W., Zhou, T., Lu, Q., Wang, X., Zhu, C., Wang, Z., & Wang, F. (2020). Dynamic fusion based federated learning for COVID-19 detection. arXiv preprint arXiv:2009.10401.
12. Liu, Y., Kang, Y., Xing, C., Chen, T., & Yang, Q. (2018). Secure federated transfer learning. arXiv preprint arXiv:1812.03337.
13. Choudhury, O., Gkoulalas-Divanis, A., Salonidis, T., Sylla, I., Park, Y., Hsu, G., & Das, A. (2019). Differential privacy-enabled federated learning for sensitive health data. arXiv preprint arXiv:1910.02578.
14. Li, W., Milletarì, F., Xu, D., Rieke, N., Hancox, J., Zhu, W., ... & Feng, A. (2019, October). Privacy-preserving federated brain tumour segmentation. In International Workshop on Machine Learning in Medical Imaging (pp. 133-141). Springer, Cham.
15. Yang, J., Shi, R., & Ni, B. (2020). MedMNIST Classification Decathlon: A Lightweight AutoML Benchmark for Medical Image Analysis. arXiv preprint arXiv:2010.14925.
16. Mironov, I., Talwar, K., & Zhang, L. (2019). R\'enyi Differential Privacy of the Sampled Gaussian Mechanism. arXiv preprint arXiv:1908.10530.
Figures