2408
PU-NET: A robust phase unwrapping method for magnetic resonance imaging based on deep learning1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Synopsis
This work proposed a robust MR phase unwrapping method based on a deep-learning method. Through comparisons of MR images over the entire body, the model showed promising performances in both unwrapping errors and computation times. Therefore, it has promise in applications that use MR phase information.
Introduction
Phase unwrapping is an important signal processing technique for many MR applications. Many practical methods have been proposed to solve this problem [1-3]. We proposed a deep MR phase unwrapping method based on an unsupervised learning network in this work. The performance of the proposed method was validated in several MR images with promising improved results compared to previously proposed PhaseNet [3].Method
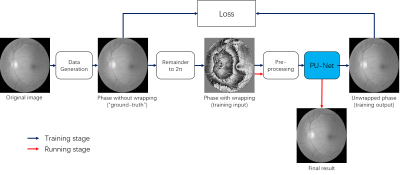
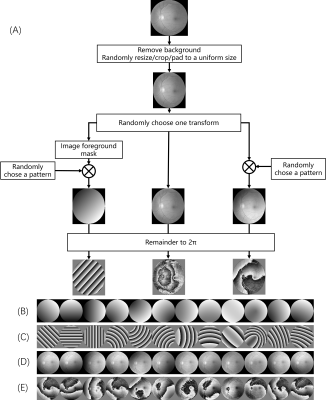
Figure 1 shows the general architecture of our proposed network. Although the unsupervised learning architecture is similar to the previously proposed PhaseNet [3], our network mainly differs in three aspects.First, instead of using randomly mixed Gaussian data, the training data were generated based on the medical image dataset [3] with several predefined patterns, as shown in Fig. 2. The original images were firstly masked out of their background and randomly resized/cropped/padded to a uniform size to facilitate training. The images were then transformed by either of the three ways. Left: extract the image foreground and generate a binary mask image with the image background set to 0, and image foreground to a random number. The mask image is multiplied with one of the predefined patterns from (B). Mid: keep the image unchanged. Right: multiply the original image with one of the predefined patterns from (B) directly. Finally, the images were taken the remainder to 2π.
Second, instead of using the wrapped phase image as the input of network directly, the wrapped phase image was underwent several pre-processing steps to avoid the interference from noisy pixels. The wrapping boundary and wrapping count estimated from the pre-processing steps, along with the original wrapped phase image were used as input of the network, as shown in Fig.3. The wrapping boundary pixels are identified in both X and Y directions by detecting whether the phase change to its neighborhood is larger than predefined thresholds. The wrapping count was estimated from the wrapping boundary information using the geometric center of image as reference point.
Finally, the model was trained by the U-net using a regression model instead of classification model used by PhaseNet.
To emphasize the importance of our proposed data generation and pre-processing steps, we trained another three networks for comparison. First, the original PhaseNet using the same training method described in[4] where the randomly mixed Gaussian images were used to generate training data without any pre-processing steps. Second, PhaseNet2.0 trained by the same training data as PU-Net but without any pre-processing steps (PhaseNet+). Third, PU-Net trained by the proposed training data but without any pre-processing steps (P0-Net).
The performance of the proposed method (PU-Net) was evaluated using various datasets by comparing it with these three networks. Some MR data were extracted from the ISMRM 2012 Challenge [5] and the Quantitative Susceptibility Mapping 2016 Challenge[6] . The other MR data were acquired from volunteers who gave informed consent and were under the approval of our institutional review board. The MR scans were performed in a 3.0 T MR system (uMR 790, Shanghai United Imaging Healthcare, Shanghai, China).
To avoid the chemical shift-induced phase change at the tissue interface, phase unwrapping was applied only to the phase factor images extracted from the multiple echo data using fat water separation method [7] .
Reults
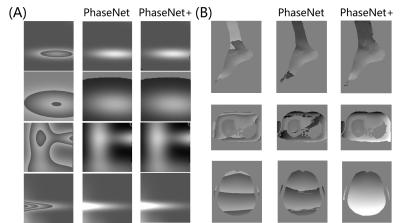
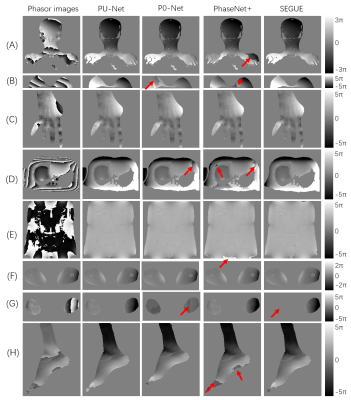
PU-Net was compared to the original PhaseNet[4] in both synthetic mixed Gaussian functions and real MR data. As shown in Fig.4, both PU-Net and PhaseNet showed good performance in the synthetic mixed Gaussian, while PhaseNet failed in several MR datasets.PU-Net was then compared to PhaseNet, PhaseNet+ and P0-Net in various anatomical images all over the whole body. The results shown in Fig. 5 demonstrate that the superior performance of PU-Net to other three networks. PU-Net was also compared to SEGUE, and achieved comparative performance to SEGUE, but the averaged computation times on the QSM 2016 Challenge were 4.87 s for PU-Net, and 18.30s for SEGUE.
Discussion and conclusions
This work proposed a robust MR phase unwrapping method based on a deep-learning method. Through comparisons of MR phase-factor images over the entire body, the model showed promising performances in both unwrapping errors and computation times. Therefore, it has promise in applications that use MR phase information.Acknowledgements
No acknowledgement found.References
[1] M. Jenkinson, “Fast, automated, N-dimensional phase-unwrapping algorithm,” Magn Reson Med, vol. 49, no. 1, pp. 193-7, Jan, 2003.
[2] A. Karsa, and K. Shmueli, “SEGUE: A Speedy rEgion-Growing Algorithm for Unwrapping Estimated Phase,” IEEE Trans Med Imaging, vol. 38, no. 6, pp. 1347-1357, Jun, 2019.
[3] "ODIR2019," https://odir2019.grand-challenge.org/.
[4] G. E. Spoorthi, S. Gorthi, and R. K. S. S. J. I. S. P. L. Gorthi, “PhaseNet: A Deep Convolutional Neural Network for Two-Dimensional Phase Unwrapping,” vol. 26, no. 1, pp. 54-58, 2018.
[5] H. H. Hu, P. B?Rnert, D. Hernando, P. Kellman, J. Ma, S. Reeder, and C. J. M. R. i. M. Sirlin, “ISMRM workshop on fat–water separation: Insights, applications and progress in MRI,” vol. 68, no. 2, pp. 378---388, 2012.
[6] "QSM RECONSTRUCTION CHALLENGE 1.0 (GRAZ 2016)," http://www.neuroimaging.at/pages/qsm.php.
[7] H. Peng, C. Zou, C. Cheng, C. Tie, Y. Qiao, Q. Wan, J. Lv, Q. He, D. Liang, and X. J. M. R. i. M. Liu, “Fat‐water separation based on Transition REgion Extraction (TREE),” 2019, 82(1): 436-448
Figures