2377
UPSS - Unsupervised perivascular spaces segmentation method with salient guidance of frangi filter1USC Stevens Neuroimaging and Informatics Institute, University of Southern California, Los Angeles, CA, United States
Synopsis
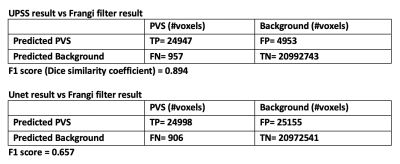
Automatic perivascular spaces (PVS) segmentation from magnetic resonance images enables whole brain morphometric assessment of glia-lymphatic network. While conventional deep learning segmentations are shown to produce reliable results, they heavily rely on the presence of ground truth for training. In this work we introduce an unsupervised learning method for PVS segmentation, by combining a rule-based image processing approach with a deep learning algorithm. The experiment results showed that the proposed method increased segmentation accuracy by effectively increasing the true positive rate compared to the rule-based method and decreasing false positive rate compared to the deep learning method.
Introduction
Perivascular spaces (PVS) attracted research attention over the recent years due to the role PVS plays in brain homeostasis, thus, effective and reliable automatic PVS segmentation methods are of high clinical and research value. Different methods have been developed using filter-based image processing algorithm like [1] or using deep supervised learning algorithm like [2]. Both methods have their own advantages and disadvantages. Normally supervised learning methods can automatically learn the segmentation process with long training time and short inference time, but these techniques are dependent on the presence of manually annotated ground truth, which is costly to acquire. The filter-based methods however use pre-defined rules, and therefore their inference time are often time consuming and their segmentation accuracy could be affected by the accuracy of the regional parcellation.In order to benefit from the pros of both filter-based and deep learning methods, here we propose a hybrid algorithm called UPSS – unsupervised perivascular spaces segmentation method with salient guidance of Frangi filter. UPSS is an unsupervised deep learning algorithm which benefits from the salient guidance of Frangi filter result, providing superior segmentation results compared to filter-based or deep learning methods.
Methods
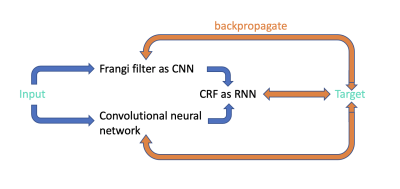
The network structure of the proposed hybrid approach is shown in Figure 1. Frangi filter [3] extracts the tubular structure probability map from enhanced perivascular space contrast image [1], which plays a role of salient post-processing guidance for convolutional neural network (CNN) [4] to reduce false positive and false negative segmentation prediction. Conditional random fields (CRF) as recurrent neural networks [5] efficiently extends post-processing method into one pass training manner instead of applying CRF inference as a post-processing step disconnected from the neural network training process. The CNN in UPSS is an encoder-decoder network with a similar structure as Unet [6].Frangi filter is also modified to CNN with fixed gaussian kernels. According to the equation 13 of Frangi filter [3], there are three pre-defined parameters α, β and c. By modifying frangi filter to CNN, parameters α, β and c can be trained automatically during the UPSS training step.
We followed the exact same CRF as RNN algorithm, except for the pairwise energy components computation. There are two components for fully connected pairwise CRF model energy function [5]: unary energy components $$$ψ_{u}$$$ and pairwise energy components $$$ψ_{p}$$$. $$$ψ_{u}$$$ is the output from Unet and $$$ψ_{p}$$$ is computed on Frangi filter results as equation 2 in [5]. We introduced self-attention (SA) [7] to compute as follows to improve network’s efficiency and accuracy,
$$K^{m}(f_{i},f_{j})=ω^{1}*SA(a_{i},a_{j})+ω^{2}*SA(p_{i},p_{j})$$
where $$$a_{i}$$$ is the appearance feature vector and $$$p_{i}$$$ is the spatial vector. $$$ω^{1}$$$ and $$$ω^{2}$$$ are two trainable parameters.
Forward-backward process is conducted during the network training. At forward stage, input goes through Frangi filter and CNN in parallel and their results gather into CRF with 8 recurrent units to get output. Then the weighted cross entropy loss will be calculated between target and output, between target and Frangi filter results and between target and CNN results. At backward stage, the three losses will backpropagate through the whole network and gradients will be calculated for each training parameters in the network and parameters will be updated accordingly.
We used human connectome project data [8]. Pre-processing and Frangi filter based PVS segmentation were done as explained in [1, 8]. We then trained our proposed network using T1w/T2w images as the input for the proposed unsupervised hybrid CNN.
Results and discussion
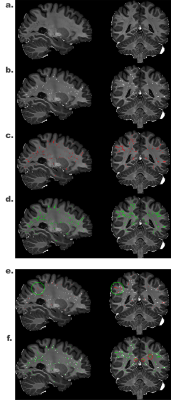
UPSS follows unsupervised learning manner because the target we used in the experiment is the result from the Frangi filter.As shown in Figure 2 e., Frangi filter binary map result are overlaid on the UPSS result. Results are highly similar except that the PVS at the most superficial part of the white matter were not segmented by the Frangi filter technique because of the imperfect white matter mask. The superficial white matter PVS were however correctly segmented by UPSS (red circle in Figure 2 e.). This improved sensitivity (true positive rate) highlights the superiority of the hybrid technique over the rule-based approach.
In Figure 2 f., Frangi filter PVS mask is overlaid on the Unet result. It can be seen that the Unet alone generated false positives PVS (green circle in Figure 2 f.). This suggests that while Unet increased the sensitivity, it also increased the false positive rate. The confusion matrix is shown in Figure 3.
Conclusion
Here we proposed UPSS, which is an end-to-end trainable network that jointly learns the parameters of the frangi filter, CNN and CRF and in one unified deep neural network. Additionally, modified CNN frangi filter with fixed gaussian kernels and incorporated CRF as RNN and novel three-path parallel backpropagation make UPSS training efficient.UPSS is an unsupervised learning method that benefits from both CNN and filter-based segmentation method. Using frangi filter generated probability map as salient map for CRF, UPSS is able to alleviate the high false positive rate of the CNN. By incorporating neural network algorithm into UPSS, we were able to resolve the PVS at the boundary of white and gray matter.
Acknowledgements
The research reported in this publication was supported by the National Institute of Mental Health of the NIH under Award Number RF1MH123223.References
[1] F. Sepehrband et al., “Image processing approaches to enhance perivascular space visibility and quantification using MRI,” Sci. Rep., vol. 9, no. 1, pp. 1–12, 2019, doi: 10.1038/s41598-019-48910-x.
[2] P. Boutinaud et al., “3D segmentation of perivascular spaces on T1-weighted 3 Tesla MR images with a convolutional autoencoder and a U-shaped neural network * Corresponding author : Marc Joliot,” bioRxiv, pp. 1–31, 2020.
[3] A. F. Frangi, W. J. Niessen, K. L. Vincken, and M. A. Viergever, “Multiscale vessel enhancement filtering,” Int. Conf. Med. image Comput. Comput. Interv. Springer, Berlin, Heidelberg, vol. 1496, pp. 130–137, 1998, doi: 10.1007/bfb0056195.
[4] Y. LeCun, P. Haffner, L. Bottou, and Y. Bengio, “Object recognition with gradient-based learning,” Shape, contour Group. Comput. vision. Springer, Berlin, Heidelb., vol. 1681, pp. 319–345, 1999, doi: 10.1007/3-540-46805-6_19.
[5] S. Zheng et al., “Conditional random fields as recurrent neural networks,” Proc. IEEE Int. Conf. Comput. Vis., vol. 2015 Inter, pp. 1529–1537, 2015, doi: 10.1109/ICCV.2015.179.
[6] O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 2015, vol. 9351, pp. 234–241, doi: 10.1007/978-3-319-24574-4_28.
[7] A. Vaswani et al., “Attention is all you need,” Adv. Neural Inf. Process. Syst., vol. 2017-Decem, no. Nips, pp. 5999–6009, 2017.
[8] D. C. Van Essen et al., “The Human Connectome Project: A data acquisition perspective,” Neuroimage, vol. 62, no. 4, pp. 2222–2231, 2012, doi: 10.1016/j.neuroimage.2012.02.018.
Figures