2334
The effects of different manual segmentation on High-Resolution T2WI Based-Radiomics in the Preoperative T Staging of Rectal Cancer1Changhai Hospital, Shanghai, China, 2Huiying Medical Technology Co., Ltd., Beijing, China
Synopsis
Manual delineation of volume of interest (VOI) is widely used in current radiomics analysis, suffering from high variability. The purpose of our study was to investigate the effects of delineation of VOIs on radiomics analysis for the preoperative T staging based on high-resolution T2WI. The result demonstrated that differences in delineation of VOIs affected radiomics analysis, the minimum contour method has better stability in extracting features and the maximum contour method has better diagnostic efficiency in patients with rectal cancer.
Introduction
Recent studies have shown that radiomics analysis has important values in identifying tumor heterogeneity and can add a further dimension to the predictive power of imaging. Segmentation of lesions occupy an important role, as the volume of interest (VOI) is directly used to extract radiomics features. The goal of this study was to explore the influence of different manual segmentation methods of rectal cancer lesions on the stability of feature extraction and the diagnostic efficiency of preoperative T staging based on high-resolution T2WI.Methods
The data of rectal cancer patients confirmed by postoperative pathology and examined by 3.0T MR T2 weighted imaging (T2WI) before surgery in our hospital from January 2017 to December 2019 were analyzed retrospectively, and included in the training set and test set respectively in chronological order. According to the pathological results, stage T1-2 patients were classified as a group of non-breakthrough the muscularis propria layer, while stage T3-4 patients were classified as the breakthrough group. Two different methods, minimum contour method (Model 1) and maximum contour method (Model 2), were used to manually segment the volume of interest of lesions (VOI). Intraclass correlation coefficient (ICC) of all features was calculated and compared. The features with ICC greater than 0.8 are selected, then least absolute shrinkage and selection operator (LASSO) algorithm was used for dimension reduction, and the features were selected which were valuable for pathological T staging. The machine learning model of multilayer perceptron (MLP) was established in the training set and verified in the test set, and receiver operating characteristic curves (ROC) of the two methods were drawn, respectively calculated the area under the curve (AUC), then compared the difference with DeLong test.Results
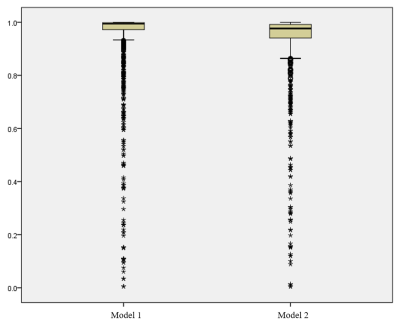
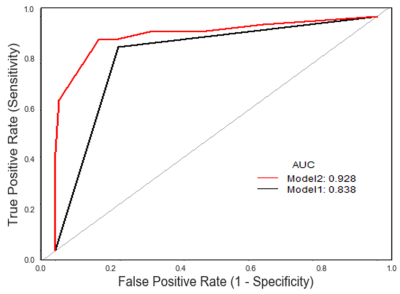
A total of 317 patients were included, including 152 cases in training set and 165 cases in test set. The median ICC of Model 1 and Model 2 was 0.994 and 0.977 respectively, and the number of features with ICC less than 0.8 was 121 (8.59%) and 136 (9.65%), respectively (Fig.1), with significant difference between the two groups (P<0.001). In the test set, the AUC (95%CI) of Model 1 was 0.838 (0.911-0.999), and the AUC (95%CI) of Model 2 was 0.928 (0.931-1.000)(Fig.2). There was significant difference between the two groups (P=0.036).Discussion and Conclusion
Our study demonstrated that the two different methods of segmentation in high-resolution T2WI based-MLP model have good diagnostic value for preoperative T staging of rectal cancer, among which the minimum contour method has better stability in extracting features and the maximum contour method has better diagnostic efficiency.Acknowledgements
NAReferences
[1] Robert JG, Paul EK, Hedvig H, et al. Radiomics: image are more than pictures, they are data[J]. Radiology. 2016; 278(2):563-577.
[2] Wu J, Tha KK, Xing L, et al. Radiomics and radiogenomics for precision radiotherapy[J]. J Radiat Res. 2018; 59(suppl_1): i25-i31.
[3] Horvat N, Veeraraghavan H, Khan M, et al. MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy[J]. Radiology. 2018; 287(3):833-843.
[4] Zhang X, Zhong L, Zhang B, et al. The effects of volume of interest delineation on MRI-based radiomics analysis: evaluation with two disease groups[J]. Cancer Imaging. 2019;19(1):89.