2333
Deep Learning Segmentation of Rectal Cancer on MRI1Department for Diagnostic Physics, Oslo University Hospital, Oslo, Norway, 2Faculty of Health Sciences, University of South-Eastern Norway, Drammen, Norway, 3University of Illinois at Chicago, Chicago, IL, United States, 4Norwegian University of Science and Technology, Trondheim, Norway, 5Akershus University Hospital, Lørenskog, Norway
Synopsis
Treatment of rectal cancer often requires repeated identification of the tumor volume by means of manual delineation by expert radiologists or oncologists. This is a tedious and time-consuming task, particularly with the growing use of multi-sequence 3D imaging. In this work, we have implemented a deep neural network for automatic detection and segmentation of rectal cancer. Our model demonstrates high detection and segmentation performance, equivalent to that of an expert reader, thus illustrating the potential use of deep learning-based segmentation in a clinically relevant setting.
INTRODUCTION
In 2018, rectal cancer was the 8th most common cancer type worldwide (1), with a rapidly increasing incidence rate for individuals younger than 50 years in the US and Europe (2). Accurate identification of the tumor is important for staging, treatment planning and outcome monitoring and prediction. Many of these patients are receiving preoperative chemoradiotherapy, where the treatment outcome benefits from continuous adaptation of radiation dose delivery to the precisely defined tumor volume during the radiotherapy treatment for optimal result. This is further highlighted by the recent introduction of the integrated MR-Linac which in the future will enable a plan-of-the-day approach, and also for quantitative MRI biomarker purposes for treatment evaluation and prediction. These approaches require repeated identification of the tumor volume. Manual delineation, the current gold standard, is a time and labor-intensive process, which is prone to inter- and intra-observer variations. MRI is part of the recommended clinical routine for treatment and follow-up of rectal cancer (3), with T2‐weighted MRI and diffusion weighted images (DWI) being among the commonly acquired sequences. This study aims to train and evaluate a deep learning-based model for accurate and robust automatic segmentation of rectal cancer with clinically available MR images as input.MATERIALS AND METHODS
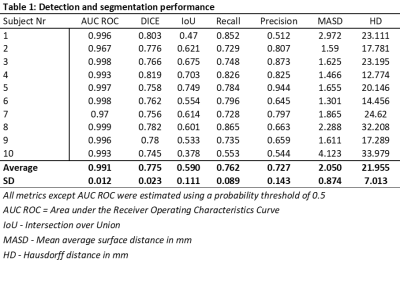
This prospective study was approved by the Institutional Review Boards and all patients provided written informed consent. A total of 110 patients with rectal cancer were included. The dataset consisted of conventional high‐resolution fast spin‐echo T2‐weighted images and DWI with a b-value of 1000 s/mm2. All images were acquired on a 1.5T Phillips Achieva system. The ground truth was established by an experienced radiologist, manually delineating whole-tumor volumes on T2‐weighted images. Neural network training was performed using a DeepLab V3 architecture with a DenseNet-101 backbone pre-trained on ImageNet. The network-output was a probability map on whether voxels represent tumor-tissue ranging from 0-1. The dataset was randomly split into 95/5/10 patients for training/validation/testing. The resulting segmentations were compared to the ground truth and evaluated by estimating the recall, precision, Intersection over Union (IoU), and Dice-score, and by using receiver operator characteristics (ROC)-curve statistics. Segmentation performance was also evaluated using the distance-based metrics mean average surface distance (MASD) and the Hausdorff distance (HD).RESULTS
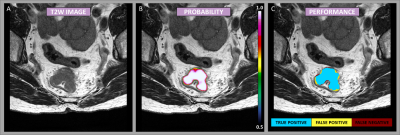
The patient cohort consisted of 73 men and 37 women, with a mean age of 65 ± 10 years (range: 41 – 88 years). Based on MRI, the adenocarcinomas were staged T2/T3/T4 with 20/37/53 cases respectively, with a median tumor volume of 23 cm3 (range: 2-234 cm3). Figure 1 shows an example case demonstrating the resulting probability map, as well as a map representing the segmentation performance as a means of true positive, false positive, and false negative, as an overlay on the T2-weighted image-series. The segmentation performance for all test cases is summarized in Table 1. The neural network showed a high voxel-wise detection accuracy, yielding an area under the ROC-curve (AUC ROC), averaged across all patients, of 0.99 ± 0.01. By using a probability threshold of 0.5 for including a voxel as a tumor, the average precision and recall, and the segmentation IoU- and Dice-score were estimated to 0.72 ± 0.14, 0.76 ± 0.09, 0.59 ± 0.11, and 0.78 ± 0.02, respectively. The distance-based measures resulted in an MASD of 2.05 ± 0.87 mm and a HD of 21.96 ± 7.01 mm.DISCUSSION
We achieved a high Dice-score when our model performance was evaluated against the manual delineation. This is in line with the inter-observer variation between manual delineations of 0.83 ± 0.13, as previously reported (4). Hence, this can be interpreted as the trained model being equally good as another expert reader. The good performance of the model is supported by the high AUC ROC values. Thus, the combination of T2-weighted and single DWI b1000 image provide sufficient information to reliably detect and segment the tumor. The results from the distance-based metrics (MASD and HD) comply with results achieved in a previous study, where a more narrow population of T3-T4 rectal tumors only were analyzed (5). It is a strength that our results are achieved in a broader population (T2-T4 tumors) with larger variation in tumor volume. The MASD and HD reported indicate that an individual user of the model may not fully agree with the automatic segmentation in all cases, depending on the exact application the results are aimed for. For some applications, such as gross tumor volume definition in radiotherapy, the user might prefer to manually modify part of the predicted segmentation, whereas for quantitative MRI biomarker purposes the segmentation may not require modification. However, it should be noted that the ultimate truth is the histologically confirmed tumor extent, and that there is an underlying uncertainty when deep learning models are trained on the manual expert contour and not the histology.CONCLUSION
Our deep-learning model provides the ability to segment the tumor volume in rectal cancer with high performance. The model has the potential to free up resources and allow for more consistent tumor volume identification in the clinic. In addition, it increases the feasibility of implementing the use of quantitative image biomarkers and adaptive radiotherapy strategies like a plan-of-the-day approach.Acknowledgements
No acknowledgement found.References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Keller DS, Berho M, Perez RO, Wexner SD, Chand M. The multidisciplinary management of rectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2020:1–16.
3. Haak HE, Maas M, Trebeschi S, Beets-Tan RG. Modern MR Imaging Technology in Rectal Cancer; There Is More Than Meets the Eye. Front. Oncol. 2020;10:2037.
4. Trebeschi S, van Griethuysen JJ, Lambregts DM, et all. Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci. Rep. 2017;7(1):1–9.
5. Wang J, Lu J, Qin G, et al. Technical Note: A deep learning‐based autosegmentation of rectal tumors in MR images. Med. Phys. 2018;45(6):2560-2564.
Figures