2269
3D Liver T1 Quantification using Interleaved Look-Locker Acquisition with T2 Preparation Pulse Sequence (3D-QALAS): Comparison with 2D-MOLLI1Imaging Research Center, Department of Radiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2MR Clinical Science, Philips, Cincinnati, OH, United States
Synopsis
Hepatic T1 relaxation values have been shown to correlate with hepatic fibrosis. The established 2D T1 mapping techniques require multiple breath-holds to quantify whole liver T1. 3D whole liver T1 quantification in a single breath-hold using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) correlates very strongly (r=0.95) with T1 values obtained with a 2D Modified Look-Locker Acquisition (2D-MOLLI). However, 3D-QALAS underestimated T1 significantly (p<0.0001) compared to 2D-MOLLI with a mean bias of 92.5 ms (14.2%). 3D-QALAS has potential to measure T1 of the whole liver in a single breath hold, while simultaneously providing T2 relaxation values.
Introduction
Quantification of the longitudinal (T1) relaxation time in the liver has been shown to provide important information in liver diagnostics [1,2]. The most accurate method of measuring hepatic T1 relaxation time is through the acquisition of a series of inversion recovery (IR) radio frequency (RF) pulse images each acquired independently at a different inversion time. However, this approach is impractical, especially in children, as it requires multiple breath-holds [3]. A two-dimensional Modified Look-locker inversion recovery (2D-MOLLI) sequence has been shown to measure hepatic T1 at a single anatomic level in a single 11 second breath-hold with good reproducibility in adults [4]. Although a few recent studies have reported hepatic T1 mapping in children [5] using a breath hold MOLLI technique, clinical adoption would be facilitated with improved speed and coverage for patients with limited ability to perform consistent breath-holds. The purpose of this study is to compare whole liver T1 quantification in a single breath-hold using an three-dimensional interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) to measurements obtained using conventional 2D Modified Look-Locker acquisition (2D-MOLLI).Methods
In this prospective HIPPA compliant IRB approved study, 19 volunteers (mean age 35.9±12.8 years, range 16-62 years, 7 males) underwent liver MR imaging with written informed consent. All imaging was performed on a Philips Ingenia 1.5T scanner (Best, The Netherlands), 28-channel (16 anterior, 12 posterior) torso coil. Scanner simulated electrocardiographic (ECG) signal of 60 beats per minute was used to trigger both the 2D-MOLLI [6] and 3D-QALAS [7] sequences. 3D-QALAS (FOV= 400x320x150 mm, acquired resolution=2.4x2.4x10 mm, 15 slices, reconstructed voxel size = 1x1x8 mm, SENSE R= 2x1.5, elliptical k-space shutter 75%) used an RF spoiled fast gradient echo acquisition schemes (TR/TE/FA=3.8/1.9/4o, echo train length = 150) to acquire transverse slices covering the entire liver. Following the first ECG trigger, a non-selective T2-preparation RF pulse (90-180-180-90, TE=100ms) was applied followed by the first data acquisition period. An IR RF pulse was then applied upon the 2nd ECG trigger, followed by 4 additional data acquisition periods, in each of the next 4 consecutive ECG intervals. This acquisition scheme is repeated three times to sample k-space, resulting in a breath hold of 15 second. T1 relaxation times were estimated from the five measurements by simulations of the longitudinal magnetizations Mz [3].2D-MOLLI [4] acquisitions (FOV= 400x320x50 mm, acquired resolution=2.3x2.3x10 mm, reconstructed voxel size = 1x1x8 mm, SENSE R=2) were performed, at 4 transverse levels through the mid-liver using 4 consecutive breath-holds of 11 second. The 2D-MOLLI acquisition used 5s(3s)3s acquisition scheme, where a non-selective inversion pulse is applied on 1st ECG trigger followed by data acquisitions in 5 consecutive heart beats, pause of 3 heart beats, followed by another non-selective inversion pulse and data acquisition for 3 consecutive heart beats; this resulted in an 11s breath-hold per slice. Exponential curve fitting is used to compute T1 relaxation times.
For measurement of liver T1, a single freehand region of interest (ROI) was drawn on each of the four 2D-MOLLI T1 maps to encompass as much of the right hepatic lobe as possible while avoiding the liver capsule, large blood vessels, dilated bile ducts, and areas of artifact. Matching ROIs were drawn on four 3D-QALAS T1 maps at corresponding slice locations. The mean T1 value for all four slices was computed for individual subject. Pearson correlation and Bland-Altman analyses were used to assess agreement. A p-value <0.05 was considered significant for all inference testing, and 95% confidence intervals were calculated as appropriate.
Results
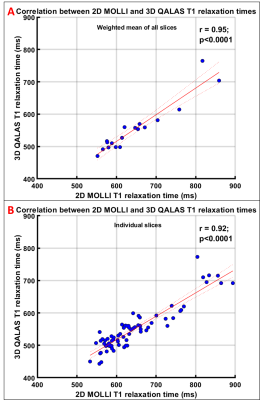
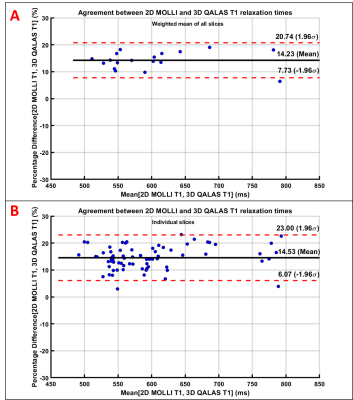
Motion artifacts were observed in 6 subjects in 3D-QALAS, one subject with severe artifacts was excluded from data analysis. In 18 subjects, with mean age of 37.3±12.6 years (range: 16-62 years; 7 males), the mean hepatic T1 values estimated by 3D-QALAS (554.67 ± 75.82 ms) and 2D-MOLLI (647.15 ± 87.31 ms) showed very strong correlation (r=0.95; p<0.0001) (Fig 1). The T1 values estimated with 3D-QALAS were significantly (p<0.0001) slower than those obtained with 2D-MOLLI with a mean bias of 92.5 ms (14.2%) with 95% confidence interval limits of 36.8 to 148.2 ms (Fig 2).Discussion
T1 relaxation values obtained by 3D-QALAS for entire liver in a single breath-hold of 15 s correlate very strongly (r= 0.95) with T1 values obtained by 2D-MOLLI, underestimating T1 significantly (p<0.0001) with a mean bias of 92.5 ms (14.2%). 3D-QALAS has the potential to allow measurement of liver T1 over a much larger area/volume of liver than 2D MOLLI. While whole liver measurement requires a 15s breath-hold, the breath-hold could be shortened by acquiring less coverage, thereby allowing an examination with fewer breath holds of shorter duration than could be achieved with a 2D MOLLI approach. Further, while not assessed as part of the current study, the 3D-QALAS sequence has the additional potential advantage of providing additional anatomic (T1-weighted, PD-weighted, T2-weighed inversion recovery) and quantitative (T2 relaxation mapping) imaging. A limitation of this study was that it included a limited number of participants.Conclusion
3D-QALAS has potential to estimate T1 of the whole liver in a single breath hold, while simultaneously estimating T2 relaxation values.Acknowledgements
No acknowledgement found.References
[1] Heye, T. et al. MR relaxometry of the liver: significant elevation of T1 relaxation time in patients with liver cirrhosis. Eur. Radiol. 2012: 22, 1224–1232.
[2] Sheng, R. F. et al. Assessment of liver fibrosis using T1 mapping on Gd-EOB-DTPA-enhanced magnetic resonance. Dig. Liver Dis. 2017: 49, 789–795.
[3] Taylor AJ. et al. T1 mapping: basic techniques and clinical applications. JACC. Cardiovasc. Imaging 2016:9, 67-81.
[4] Yoon JH. et al. Quantitative assessment of hepatic function: modified look-locker inversion recovery (MOLLI) sequence for T1 mapping on Gd-EOB-DTPA-enhanced liver MR imaging. Eur. Radiol. 2016:26, 1775-1782.
[5] Gilligan LA. et al. Magnetic resonance imaging T1 relaxation times for the liver, pancreas and spleen in healthy children at 1.5 and 3 tesla. Pediatr. Radiol. 2019:49, 1018-1024.
[6] Messroghli DR. et al. Modified look-locker inversion recovery (MOLLI) for high-resolution T 1 mapping of the heart. Magn Reson Med. 2004:52, 141-146.
[7] Kvernby S. et al. Simultaneous three-dimensional myocardial T1 and T2 mapping in one breath hold with 3D-QALAS. JCMR 2014:16, 102.