2259
A semi-supervised graph convolutional network for early prediction of motor impairments in very preterm infants using brain connectome1Imaging Research Center, Department of Radiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Department of Electrical Engineering and Computer Science, University of Cincinnati, Cincinnati, OH, United States, 3Deep MRI Imaging Inc., Lewes, DE, United States, 4The Perinatal Institute and Section of Neonatology, Perinatal and Pulmonary Biology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 5Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, United States
Synopsis
About 32−42% of very preterm infants develop minor motor impairments around the world. Unfortunately, large MRI datasets with clinical outcome annotations/labels are typically unavailable, especially in neonates. To address this challenge of limited training data, we developed a semi-supervised graph convolutional network model to utilize both labeled and unlabeled data during model training to predict motor impairments at 2 years corrected age using brain structural connectome derived from diffusion MRI obtained at term-equivalent age in very preterm infants. The proposed model was able to identify infants with motor impairments with an accuracy of 68.1% and an AUC of 0.67.
Introduction
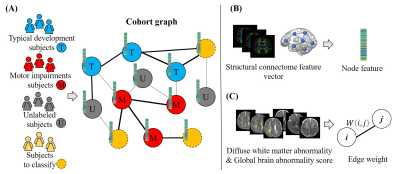
About 32−42% of very preterm infants (VPIs; 32 weeks’ gestational age) develop minor motor impairments around the world.1 Motor impairments, especially in infants without moderate or severe brain injuries, typically cannot be diagnosed until 1-2 years of age. The application of deep learning models to diffusion MRI (dMRI) has been demonstrated to make feasible predictions of motor impairments soon after birth in VPIs.2,3 These models were typically trained in a supervised learning manner, which requires a large number of labeled training data. To address the challenge of limited labeled training data, we developed a semi-supervised graph convolutional network (GCN) model to utilize both labeled and unlabeled training data.The semi-supervised GCN model4 was originally proposed to model graph-structured data, and has been applied to the diagnosis of autism spectrum disorder and Alzheimer's disease.5 In this work, we proposed to develop a semi-supervised GCN model to predict motor impairments at 2 years corrected age (CA) using brain structural connectome derived from dMRI obtained at term-equivalent age in VPIs. Specifically, we construct a cohort graph, in which each node represents a subject and each weighted edge represents inter-subject similarity. The node feature is a structural connectome feature vector. The edge weight is calculated based on the diffuse white matter abnormality (DWMA) volume and global brain abnormality (GBA) score derived from anatomical MRI images.
Methods
Subjects and MRI data acquisitionWe enrolled 260 VPIs from five Greater Cincinnati area NICUs. The Cincinnati Children’s Hospital Institutional Review Board approved this study. Parents or guardians of infants gave written informed consent before enrollment. All subjects were imaged during unsedated sleep on a 3T Philips Ingenia scanner with a 32-channel receiver head coil. Acquisition parameters--dMRI: repetition time (TR) 6972 ms, echo time (TE) 88 ms, flip angle (FA) 90°, resolution 2 × 2 × 2 mm3, 36 directions, b value = 800 s/mm2. Axial T2-weighted images: TR 18567 ms, TE 166 ms, FA 90°, resolution 1.0 × 1.0 × 1.0 mm3. 3D T1-weighted images: TE 3.4 ms, TR 7.3 ms, FA 11°, resolution 1.0 × 1.0 × 1.0 mm3.
MRI data processing
The final cohort contains 224 VPIs after image quality control. For each subject, we quantified whole-brain normalized DWMA on T2-weighted images using our published method.6 GBA score was obtained using a standardized scoring system7 based on T1- and T2-weighted MRI images. The dMRI data were preprocessed and the structural brain connectome was constructed using our prior published method.8
Graph convolutional networks
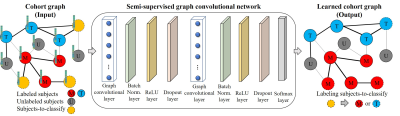
The input of the proposed GCN model is a cohort graph (Figure 1), where each node represents a subject and the node feature is the structural connectome feature vector (i.e., a vector of 4005, representing each pair of 90 regions of interests); and each weighted edge is the inter-subject similarity, whose weight between subjects i and j is calculated as $$$ W(i,j)=e^{-(|\gamma(i)-\gamma(j)|+|\delta(i)-\delta(j)|)} $$$, where $$$ \gamma $$$ is the normalized DWMA volume and $$$ \delta $$$ is GBA score. The cohort graph was built using both labeled and unlabeled data. The GCN model consisted of two graph convolutional layers, each of which was followed by a batch normalization layer, a rectified linear unit activation layer, and a dropout layer. (Figure 2) The model was then trained to assign labels (i.e., motor impairment vs. typical development) to subjects-to-classify. We used a weighted binary cross-entropy loss function to mitigate the imbalanced dataset issue. The Adam algorithm9 was applied to train the model with a learning rate of 0.01 and an epoch of 2000. We applied T-test (p-value < 0.05) on training data to reduce the dimension of the structural connectome feature vector. We evaluated the model using accuracy, balanced accuracy, sensitivity, specificity, and area under the curve (AUC). A 5-fold cross-validation was applied and repeated 10 times to evaluate model reproducibility.
Results
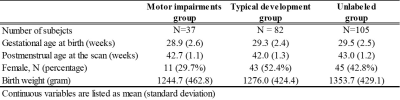
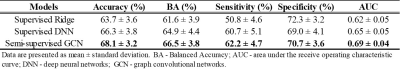
Out of 224, 119 VPIs undergone standardized Bayley Scales of Infant and Toddler Development III test at 2 years CA. A cutoff value of 85 was utilized to dichotomize the cohort into motor impairments vs. typical development groups. The baseline demographics of the cohort are listed in Table 1.Table 2 shows that the semi-supervised GCN model identified subjects with motor impairments with an accuracy of 68.1% and an AUC of 0.69. It achieved better prediction performance than the supervised deep neural network with an increase of 1.8% on accuracy (p=0.035) and 0.04 on AUC (p<0.001). The proposed model also outperformed the traditional machine learning Ridge model on accuracy (p=0.004) and AUC (p<0.001).
Discussion and Conclusion
Early identification of VPIs with motor impairments is critical for improving their quality of life. We developed a semi-supervised GCN model for the early prediction of motor impairments at 2 years in VPIs using both labeled and unlabeled training data. The cohort graph construction is the key, since an inappropriately constructed graph may hinder the model training. The main limitation of current work is that semi-supervised GCN belongs to transductive learning, which requires training data to be available when the model infers labels for new samples. We demonstrated that the proposed model outperformed two peer supervised models. Our future directions include the optimization of graph construction to further improve the prediction performance.Acknowledgements
This study was supported by the National Institutes of Health grants R21-HD094085, R01-NS094200, R01-NS096037, R01-EB029944, and a Trustee grant from Cincinnati Children’s Hospital Medical Center.References
1. Williams J, Lee KJ, Anderson PJ. Prevalence of motor-skill impairment in preterm children who do not develop cerebral palsy: a systematic review. Developmental medicine and child neurology. 2010;52(3):232-237.
2. Saha S, Pagnozzi A, Bourgeat P, et al. Predicting motor outcome in preterm infants from very early brain diffusion MRI using a deep learning convolutional neural network (CNN) model. Neuroimage. 2020;215:116807.
3. Kawahara J, Brown CJ, Miller SP, et al. BrainNetCNN: Convolutional neural networks for brain networks; towards predicting neurodevelopment. Neuroimage. 2017;146:1038-1049.
4. Kipf TN, Welling M. Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:02907. 2016.
5. Parisot S, Ktena SI, Ferrante E, et al. Disease prediction using graph convolutional networks: Application to Autism Spectrum Disorder and Alzheimer’s disease. Medical image analysis. 2018;48:117-130.
6. He L, Parikh NA. Atlas-guided quantification of white matter signal abnormalities on term-equivalent age MRI in very preterm infants: findings predict language and cognitive development at two years of age. PloS one. 2013;8(12):e85475.
7. Kidokoro H, Neil JJ, Inder TE. New MR imaging assessment tool to define brain abnormalities in very preterm infants at term. AJNR American journal of neuroradiology. 2013;34(11):2208-2214.
8. Chen M, Li H, Wang J, et al. Early Prediction of Cognitive Deficit in Very Preterm Infants Using Brain Structural Connectome With Transfer Learning Enhanced Deep Convolutional Neural Networks. Front Neurosci. 2020;14:858.
9. Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:14126980. 2014.
Figures