2206
Rapid MRSI of the Brain on 7T Using Subspace-Based Processing1Bioengineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois Urbana-Champaign, Urbana, IL, United States, 3Radiology, University of Pittsburgh, Pittsburgh, PA, United States
Synopsis
We present here a new method for rapid MRSI of the brain on 7T that combines fast rosette trajectories and subspace-based processing from the SPICE MRSI framework. Results from in vivo experiments demonstrated substantial improvement in spectral quality and SNR achieved by the subspace imaging method over the standard reconstruction of rosette MRSI data. The ability to obtain single-slice rapid MRSI of the brain with an only 1.1 min acquisition was also demonstrated using the subspace-based reconstruction. These results highlight the potentials of integrating subspace imaging and fast acquisitions to push the performance of ultrahigh-field MRSI.
INTRODUCTION
Improvements in SNR, speed and spectral quality are key motivations for developing MRSI on ultrahigh-field systems.1 In the past few years, a number of sequences and customized instrumentations have been developed for fast, high-resolution MRSI at 7 and 9.4T.2-7 While these methods have achieved excellent performance, spectral quality problems persist, particularly from nuisance lipid and/or macromolecule signals. Furthermore, better SNRs through advanced processing are highly desired in order to achieve better resolution and speed tradeoffs for fast trajectories.8-10 We present a new processing method for brain MRSI at 7T based on the recently proposed subspace imaging approach11-12 to improve the spectral quality of data from fast rosette spectroscopic imaging (RSI).10 Results from in vivo experiments showed substantial improvement in residual nuisance water/lipid signal reduction and SNR enhancement for 7T RSI data achieved by the proposed method over the standard reconstruction. We also demonstrated the capability of our subspace-based method in producing high-fidelity reconstruction from temporally undersampled RSI data, which can enable further acceleration by reducing the number of temporal interleaves. These results highlight the potentials of synergistic acquisition and processing strategies to advance ultrahigh-field MRSI.METHODS
Data from five healthy volunteers were acquired on a Siemens 7T Magnetom 8 channel pTx system with the 8x2 transceiver using two RF-shimmed B1+ distributions (homogeneous, ring) as previously described.7,13 Outer volume suppression of extracerebral tissue was performed using the B1+ ring spatial excitation. B0 shimming was performed using a high-order shim insert (Resonance Research Inc.), achieving water linewidths of 4-5Hz over a frontal-parietal slice.14 A J-refocused double-spin-echo sequence was used with TR/TE=1500/34ms (single slice, 2.2min total), which provides excellent retention of coupled spins through longer TEs.15 Spatial encoding was performed using a circular rosette trajectory with a 216x216mm2 FOV, 9mm slice thickness, 44 shots (9x9mm2 in plane voxels), two temporal interleaves, and 2500Hz spectral bandwidth. More details for the pulse sequence can be found in a concurrently submitted abstract. Coil combination was performed using an SVD-based scheme (other SNR-optimized strategies can be considered). A MP2RAGE scan was acquired to extract the spatial support of brain and extracerebral tissues for water/lipids removal (NSRM).11-12 B0 maps were also acquired for data processing.While this acquisition achieves high SNR and good lipid suppression, significant nuisance extra-cerebral signal leakage can remain and result in poor spectral quality, especially for the edge/cortical voxels. The OVS itself can also reduce the SNR at the edge through the imperfect ring distribution, thereby perturbing the intracerebral signal. With the availability of accurate B0 maps and structural images, we address the NSRM and SNR issues using subspace-based processing. Furthermore, given the spectral prior knowledge afforded by a pre-estimated subspace11,16, we demonstrate that with our subspace-based reconstruction, data acquisition can be further accelerated through less temporal interleaves. Specifically, we formulate the reconstruction problem as follows10-11
$$\{\hat{\mathbf{U}}_w,\hat{\mathbf{U}}_f,\hat{\mathbf{U}}_m,\hat{\mathbf{V}}_m\}=\arg\underset{\{\mathbf{U}_w,\mathbf{U}_f,\mathbf{U}_m,\mathbf{V}_m\}}{\min}\left\Vert\mathbf{d}-\mathcal{F}_{\Omega}\left\{\mathbf{B}\odot\left(\mathbf{M}_w\mathbf{U}_w\hat{\mathbf{V}}_w+\mathbf{M}_f\mathbf{U}_f\hat{\mathbf{V}}_f+\mathbf{U}_m\mathbf{V}_m\right)\right\}\right\Vert_2^2+\lambda_w\left\Vert \mathbf{U}_w\right\Vert_F^2+\lambda_f\left\Vert \mathbf{U}_f\right\Vert_F^2+\lambda_m\left\Vert \mathbf{D}_w\mathbf{U}_m\mathbf{V}_m\right\Vert_F^2, $$
where $$$\mathbf{d}$$$ denotes (k,t)-space data, $$$\mathcal{F}_{\Omega}$$$ is a encoding operator that maps Cartesian-grid images to the rosette data, $$$\mathbf{B}$$$ captures B0 inhomogeneity related phases, and $$$\mathbf{M}_x$$$ captures the spatial support information (1's if signal present and 0's otherwise) for water and lipids, respectively. $$$\mathbf{U}_x\mathbf{V}_x$$$ are explicit low-rank (subspace) constraints for water, lipids and metabolites, respectively. We solve this problem using a two-step approach. More specifically, we first gridded the RSI data to Cartesian grids (24x24) and performed B0 correction followed by a parametric fitting to estimate the subspaces ($$$\hat{\mathbf{V}}_w$$$, $$$\hat{\mathbf{V}}_f$$$ and $$$\hat{\mathbf{V}}_m$$$).11 We then refit the data to estimate water/lipid signals on a higher-resolution grid (64x64) and removed their contributions from the data. Finally, a joint low-rank and spatially constrained reconstruction was performed to obtain the metabolite spatial-spectral distribution.
RESULTS
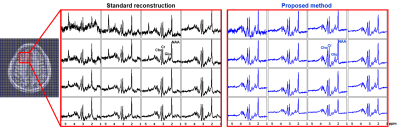
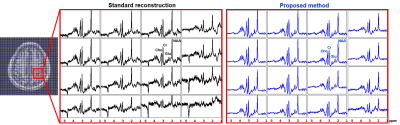
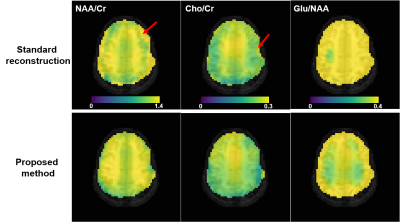
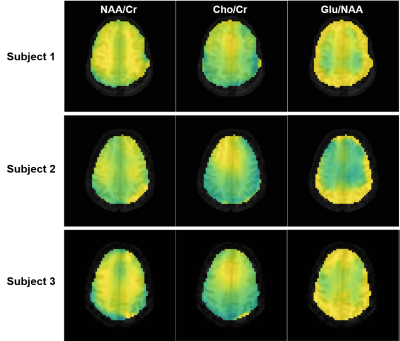
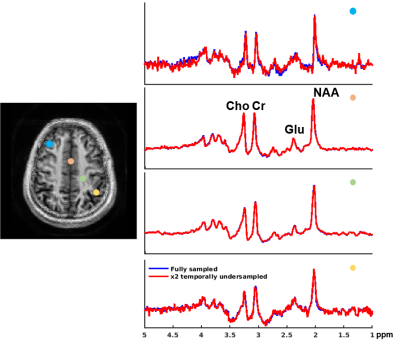
We evaluated the proposed method in different scenarios to illustrate its utility for fast 7T MRSI. First, we compare the quality of spatial-spectral reconstruction produced by our subspace method to that by the standard reconstruction (gridding+FFT+coil combination10). Figures 1 and 2 compare spatially-localized spectra. It is clear that the proposed method yielded spectra with significant reductions of lipid/water contamination and higher SNRs, especially for the edge voxels. Metabolite ratio maps (i.e., NAA/Cr, Cho/Cr and Glu/NAA) from the standard reconstruction and the proposed method are shown in Fig. 3, the latter showing better gray/white matter contrast. Metabolite maps from different subjects are shown in Fig. 4.To examine the capability of the subspace method in enabling temporal undersampling, we also performed reconstruction using only a single temporal interleave (corresponding to a 1.1min acquisition). Figure 5 compares the reconstruction from this undersampled data with the full data. Very similar spatially-resolved spectra were produced, demonstrating the capability of subspace-based reconstruction for further acceleration. Note that the subspace used in this reconstruction was pre-estimated from three RSI data with full spectral width other than the one under evaluation. More details on subspace learning can be found in Refs. [11] and [16].
CONCLUSION
We demonstrate that subspace modeling can improve the performance of fast MRSI of the brain at 7T. Further acceleration using subspace-based reconstruction can be achieved. The results obtained highlight the unique potentials of synergistic acquisition and processing strategies to push the limits of ultrahigh-field MRSI. Future work will include higher-resolution acquisitions, multislice coverage and further optimized reconstruction methods.Acknowledgements
No acknowledgement found.References
1. Otazo R, Mueller B, Ugurbil K, Wald L and Posse S. Signal-to-noise ratio and spectral linewidth improvements between 1.5 and 7 tesla in proton echoplanar spectroscopic imaging. Magn Reson Med. 2006;56:1200-1210.
2. Henning A, Fuchs A, Murdoch JB and Boesiger P. Slice‐selective FID acquisition, localized by outer volume suppression (FIDLOVS) for 1H‐MRSI of the human brain at 7 T with minimal signal loss. NMR Biomed, 2009;22:683-696.
3. Bogner W, Gruber S, Trattnig S and Chmelik M. High-resolution mapping of human brain metabolites by free induction decay 1H MRSI at 7 T. NMR Biomed, 2012;25:873-882.
4. Hangel G, Strasser B, Povazan M, et al. Ultra-high resolution brain metabolite mapping at 7 T by short-TR Hadamard-encoded FID-MRSI. NeuroImage, 2018;168:199-210.
5. Nassirpour S, Chang P and Henning A. High and ultra-high resolution metabolite mapping of the human brain using H-1 FID MRSI at 9.4T. NeuroImage, 2018;168:211-221.
6. Chadzynski GL, Bause J, Shajan G, Pohmann R, Scheffler K and Ehses P. Fast and efficient free induction decay MR spectroscopic imaging of the human brain at 9.4 Tesla. Magn Reson Med. 2017;78:1281-1295.
7. Hetherington HP, Avdievich NI, Kuznetsov AM and Pan JW. RF shimming for spectroscopic localization in the human brain at 7 T. Magn Reson Med. 2010;63:9-19.
8. Chiew M, Jiang W, Burns B, et al. Density‐weighted concentric rings k‐space trajectory for 1H magnetic resonance spectroscopic imaging at 7 T. NMR Biomed, 2018;31:e3838.
9. Moser P, Bogner W, Hingerl L, et al. Non‐Cartesian GRAPPA and coil combination using interleaved calibration data – application to concentric‐ring MRSI of the human brain at 7T. Magn Reson Med, 2019;82:1587–1603.
10. Schirda CV, Tanase C and Boada FE. Rosette spectroscopic imaging: Optimal parameters for alias‐free, high sensitivity spectroscopic imaging. J Magn Reson Imaging, 2009;29:1375-1385.
11. Lam F, Ma C, Clifford B, et al. High-resolution 1H-MRSI of the brain using SPICE: data acquisition and image reconstruction. Magn Reson Med, 2016;76:1059–1070.
12. Ma C, Lam F, Johnson CL and Liang ZP. Removal of nuisance signals from limited and sparse 1H MRSI data using a union-of-subspaces model. Magn Reson Med, 2016;75:488–497.
13. Li X, Pan JW, Avdievich NI, Hetherington H and Rispoli JV. Electromagnetic simulation of a 16-channel head transceiver at 7T using circuit-spatial optimization. Magn Reson Med. 2020. In Press.
14. Hetherington HP, Moon CH, Schwerter M, Shah NJ and Pan JW. Dynamic B0 shimming for multiband imaging using high order spherical harmonic shims. Magn Reson Med. 2021;85:531-543.
15. Pan JW, Avdievich N and Hetherington HP. J-refocused coherence transfer spectroscopic imaging at 7 T in human brain. Magn Reson Med. 2010;64:1237-46.
16. Lam F, Li Y, Guo R, Clifford B and Liang ZP. Ultrafast magnetic resonance spectroscopic imaging using SPICE with learned subspaces. Magn Reson Med. 2020;83:377-390.
Figures