2195
Ablation Studies in 3D Encoder-Decoder Networks for Brain MRI-to-PET Cerebral Blood Flow Transformation1Radiology, Stanford University, Stanford, CA, United States, 2Bioengineering, University of California, Riverside, Riverside, CA, United States, 3Neuro MR, GE Healthcare, Menlo Park, CA, United States
Synopsis
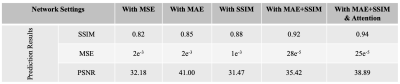
In this study, we demonstrate that an optimized 3D encoder-decoder structured convolutional neural network with attention gates can effectively integrate brain structural MRI and ASL perfusion images to produce high-quality synthetic PET CBF maps without using radiotracers. We performed experiments to evaluate different loss functions and the role of the attention mechanism. Our results showed that attention-based 3D encoder-decoder network with custom loss function produces the superior PET CBF prediction results, achieving SSIM of 0.94, MSE of 0.00025, and PSNR of 38dB.
Introduction
Cerebral blood flow (CBF) is the blood supply to the brain in a given period of time. Accurate quantification of CBF plays a crucial role in the diagnosis and assessment of cerebrovascular diseases such as Moyamoya, Stroke, and intracranial atherosclerotic steno-occlusive disease (ICSD).1 This study first presents a deep encoder-decoder neural network for predicting O15-water PET CBF maps from multi-contrast brain MRI images. We also conduct ablation studies to evaluate the performance of various components of the proposed encoder-decoder structure.Methods
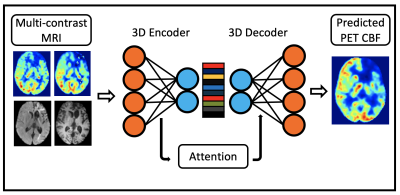
Data were acquired from 32 healthy controls and 32 patients with cerebrovascular diseases (i.e., 26 with Moyamoya and 6 with ICSD) on a 3T PET/MRI hybrid system (SIGNA, GE Healthcare) using an 8-channel head coil. The MRI scans included T1, FLAIR, single-delay arterial spin labeling (ASL), and multi-delay pseudo-continuous ASL. The structural MRI images and quantified CBF maps derived from ASL were co-registered and normalized to the Montreal Neurological Institute (MNI) brain template, and then resized to 96×96×64 voxels.We first introduce a deep encoder-decoder architecture that leverages the power of 3D fully convolutional neural network (CNN), trained end-to-end, to generate high-quality synthetic PET CBF maps from multi-contrast MRI images, as shown in Figure 1. The encoder performs a series of 3D convolution to compress the input multimodal MRI volumes into low-resolution feature maps, while the decoder is responsible for 3D deconvolution and un-pooling/up-sampling to predict pixel-wise PET CBF measurements.2 The proposed network offers a significant improvement in performance in PET CBF prediction results while optimizing the number of trainable parameters.
To further gain some insights of the improvements obtained by the proposed architecture, we conduct more additional experiments for ablation studies as listed in Figure 2, where we examine the effectiveness and contributions of different strategies used in the proposed architecture. Figure 2 shows the detailed experimental results of the ablation studies for examining several different settings. The prediction performance of the proposed 3D encoder-decoder architecture was evaluated in terms of the structural similarity index (SSIM), mean square error (MSE), and peak signal-to-noise ratio (PSNR), using 4-fold cross-validation.
Results and Discussion
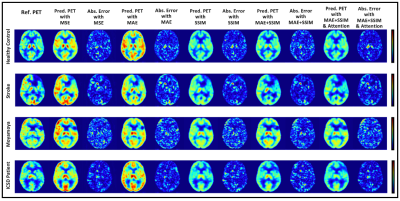
Reference CBF maps, corresponding synthetic CBF maps produced at different network settings from representative subjects are shown in Figure 3. We have experimented with different loss functions such as MSE, mean absolute error (MAE), SSIM, a weighted summation of MAE and SSIM, to design an efficient convolutional encoder-decoder architecture. We further determined the importance of attention mechanisms used by the network’s decoder for the CBF prediction solely based on a subset of the encoder feature maps. To be precise, in this work, we not only remove parts of the network but also substituting them with (more appropriate) alternative constructs.The initial results show that integrating multiple communicative MRI modalities can notably improve the prediction of PET CBF maps for healthy controls and also patients with cerebrovascular diseases. Compared to the previous CBF prediction method that achieved an average SSIM of 0.84,3 our model yields superior performance up to 0.94 SSIM.
We argue that ablations studies are a feasible method to investigate knowledge representations in encoder-decoder networks and are especially helpful to examine networks performance and reliability against structural artifacts and ghosting. The performance of different loss functions and network settings, and the incremental performance gain of each component can be seen in Figures 2 and 3. We can see steady improvement in both quantitative and qualitative CBF prediction results as we use more appropriate loss functions. Also, attention mechanisms enable the network to focus more on the relevant aspects of the input at the channel and spatial levels,2 thus yielding improved synthetic CBF map quality. On the whole, attention-based 3D encoder-decoder networks with an adequate loss function can effectively exploit channel-level and spatial-level information and efficiently produce high-quality PET CBF maps.
Conclusion
This work demonstrates that a 3D convolutional encoder-decoder network integrating multi-contrast information from brain structural MRI and ASL perfusion images can synthesize high-quality PET CBF maps. It also shows how the weighted custom loss functions can improve the PET CBF prediction results and how attention mechanisms can boost the prediction performance of encoder-decoder networks.Acknowledgements
No acknowledgement found.References
1. Bandera E, Botteri M, Minelli C, Sutton A, Abrams KR, Latronico N. Cerebral blood flow threshold of ischemic penumbra and infarct core in acute ischemic stroke: a systematic review. Stroke 2006; 37:1334-1339.
2. Cho K, Courville A, Bengio Y. Describing multimedia content using attention-based encoder-decoder networks. IEEE Trans Multimedia 2015; 17:1875-1886.
3. Guo J, Gong E, Fan AP, Goubran M, Khalighi MM, Zaharchuk G. Predicting 15O-water PET cerebral blood flow maps from multi-contrast MRI using a deep convolutional neural network with evaluation of training cohort bias. J. Cereb. Blood Flow Metab 2019; 0271678X19888123.
Figures