2188
Automatic Quantitative Analysis of Low-field Infant Brain MR Images1Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou, China, 2Suzhou Key Laboratory of Medical and Health Information Technology, Suzhou, China, 3Jinan Guoke Medical Engineering Technology Development co., Ltd., Jinan, China, 4Department of Radiology, Children’s Hospital of Soochow University, Suzhou, China, 5Department of Radiology, The First People’s Hospital of Lianyungang, Jiangsu Province, China, 6Department of Medical Imaging, Lianyungang Women and Children Hospital and Health Institute, Jiangsu Province, China, 7Jiangsu LiCi Medical Device Co., Ltd., Lianyungang, China
Synopsis
Low-field MRI is foreseeable as a safer system for infants. However, low-field MR images have lower SNR and spatial resolution as compared to high-field images, thus processing of low-field infant brain MR image is challenging. In this study, an automated image processing method that can accurately perform brain extraction, tissue segmentation, and brain labeling on low-field infant brain MR images is developed. It is also capable to automatically construct the inner, middle, and outer surfaces of the cerebral cortex and provides automatic quantitative analysis of selected region of interest, which can be a helpful tool for researchers in neuroimaging studies.
Introduction
Infant period (first 12 months) is critical in the process of brain development as it undergoes rapid physical growth and functional evolution. Particularly, the total brain volume and the area of the cortex will increase and expand greatly1. Hence, quantitative analysis of infant brain structure via volumetric and cortical measurements of the brain tissue can provide effective means to study the relationship between brain development and neurobehavioral abilities. In recent years, low-field MRI is foreseeable as a safer system for infant brain scanning but low-field images have lower SNR and spatial resolution as compared to high field images2. In addition, due to the size of the infant brain and the difficulty in differentiating gray-white tissue, processing of low-field infant brain MR images can be challenging. Although there are available adult specific brain MR image processing methods3, they are, however, not directly adapted for infant use. In this work, we proposed a low-field infant brain MR images processing method that is capable to automatically perform segmentation of the gray matter (GM), white matter (WM), cerebrospinal fluid (CSF), and the reconstruction of the cerebral cortex. In addition, anatomically-meaningful regions of interests (ROIs) can be automatically labeled and quantitative analyzed. The proposed method is tested on 0.35T infant brain MR images and is shown that the proposed method can accurately perform segmentation and quantitative determination.Method
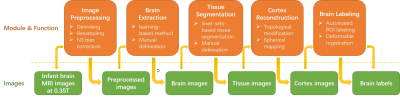
Thirteen infant brain MR images are acquired using a 0.35T dedicated neonatal-infant brain MRI system equipped with a two channels transceive RF head coil. Using a 3D-GRE sequence, T1W images are acquired (TR/TE=45 ms/15 ms, flip angle=90 deg, FOV=200 mm, slice thickness=3 mm, Matrix size=224 × 320), and T2W images are also acquired using a 2D-FSE sequence (TR/TE=4037 ms/114 ms, flip angle=90 deg, FOV=220 mm, slice thickness=6 mm, Matrix size=224 × 256). The image processing flowchart is shown in Figure 1 (consists of five major steps). The original data first undergo a series of preprocessing steps, including image de-noising, resampling of each image to a standard format (RAS coordinate, voxel size 1 mm, and volume size 256 × 256 ×256), and intensity bias correction to improve intensity homogeneity. For brain extraction, tissue segmentation, and brain labeling a combined brain analysis algorithms are implemented3-5. A learning-based meta-algorithm based on BSE and BET is adopted for infant brain extraction4. Using the learning-based method, the appropriate parameters for different infant age months and different development stages of the infant brain tissues can be trained respectively. A level-sets-based tissue segmentation algorithm is used to accurately segment the infant brain image into GM, WM, CSF, and background. In addition, due to topology error, there existed some small hole-like errors in the surface reconstruction of the GM, WM, and CSF and topology correction algorithm is used to correct these errors. An automated ROI labeling algorithm based on HAMMER registration is used5. Workstation installed with 8G memory, 32GB disk and Linux operating system (64 bit) is used for the implementation of the proposed method.Result
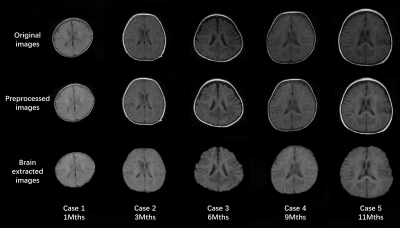
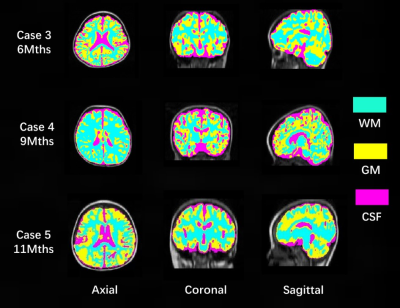
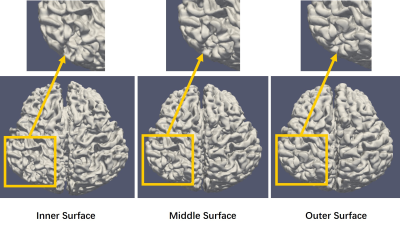
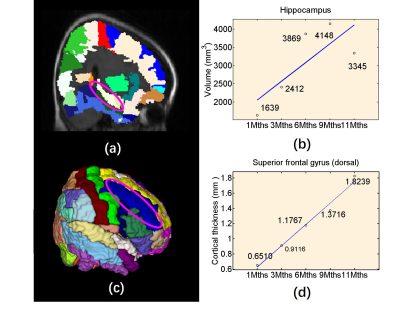
Top row of Figure 2 shows the original acquired low-field infant brain MR images at various age months. Using our processing method, middle row of Figure 2 depicted the preprocessed images and bottom row of Figure 2 shows the brain extraction results. The segmented results of the GM, WM, and CSF are shown in Figure 3, while Figure 4 displayed the reconstructed inner (GM surface), middle (surface between GM and WM), and outer layers (WM surface) of the cerebral cortex. After labeling, quantitative analysis for selected ROI can be undertaken through the voxel-wise image and surface-based image, and Figure 5 shows the quantitative determination results of the cortical volume and thickness.Discussion
In the proposed low-field infant brain MR image processing and analysis method, infant brain specific algorithms have been developed and tested. Using the acquired low-field infant brain MR images, it is shown that our method is capable to automatically extract, segment and label the infant brain with high accuracy. It is, however, noted that for neonatal (particularly <1 month) the gray-white tissue contrast is very low, hence; a secondary images (in this case a T2W images) have to be used to achieve a more accurate segmentation results. In addition, the ability to perform automatic quantitative analysis of selected ROI can help in identify quantitative changes of the anatomical regions in the early stage of the brain development, which is helpful in detecting infantile brain diseases such as autism, hypoxic-ischemic encephalopathy and brain hypoplasia.Conclusion
Development of cerebral cortex in infants is dynamic and not completely formed and the over development of sulcus and gyrus can lead to enlargement of brain volume and the expansion of cortical area. It is difficult to know how much volume is increased and how much area is expanded. Using our proposed processing and analysis method, it is possible to establish a roadmap, which can accurately reflect the dynamic changes of infant cortical surface. It has important clinical significance as it can help with the study of infant brain development mechanism and early diagnosis and intervention of neonatal and infant brain diseases.Acknowledgements
This study was supported by National Nature Science Foundation of China under Grant (61801476), Jiangsu Key Technology Research Development Program (BE2018610), Jiangsu Natural Science Foundation (BK20180221), Quancheng 5150 Project, Jinan Innovation Team (2018GXRC017), Suzhou Science and Technology Project (SS201855, SS210866, SS2019012; SS202065).References
1. Li, Gang, et al. Mapping region-specific longitudinal cortical surface expansion from birth to 2 years of age. Cerebral cortex 22.11(2013): 2724-2733.
2. Coffey, Aaron M., M. L. Truong, and E. Y. Chekmenev. "Low-field MRI can be more sensitive than high-field MRI." Journal of Magnetic Resonance 237(2013):169-174.
3. Dai, Yakang, et al. "iBEAT: a toolbox for infant brain magnetic resonance image processing." Neuroinformatics 11.2(2013): 211-225.
4. Shi, Feng, et al. "Altered structural connectivity in neonates at genetic risk for schizophrenia: A combined study using morphological and white matter networks." Neuroimage 62.3(2012): 1622-1633.
5. Shen, Dinggang, and C. Davatzikos. "HAMMER: hierarchical attribute matching mechanism for elastic registration." IEEE Transactions on Medical Imaging 21.11(2002): 1421.
Figures