2167
Automated quantitative evaluation of deep learning model for reduced gadolinium dose in contrast-enhanced brain MRI1Subtle Medical Inc, Menlo Park, CA, United States, 2Subtle Medical Inc., Menlo Park, CA, United States
Synopsis
Deep learning (DL) has recently become a promising technique to address the safety concerns for Gadolinium-based Contrast Agents (GBCAs) in MRI. Studies have shown that high quality contrast-enhanced images can be generated by DL with only a small fraction of the standard dose, and in some cases no dose at all. To build upon existing research that has heavily focused on qualitative evaluation by radiologists, this work proposes an automated quantitative evaluation scheme for the GBCA dose reduction using DL.
Introduction
Given the safety issues of GBCAs due to its prolonged retention in the human tissues [1], there is an increased need to reduce its usage without compromising image contrast and quality. A DL framework was recently proposed [2] that could generate non-inferior contrast-enhanced images using only 10% of the standard dose. Building upon this work, a generic DL model with comprehensive technical solutions was proposed [3]. To date, existing research has been focused on qualitative evaluation by radiologists to validate the efficacy of the DL models. However, this step is time consuming, especifically for a large patient cohort. To automate the evaluation of diagnostic performance, DL-based tumor segmentation algorithms are good and accurate alternatives [4]. This work proposes an automated DL-based quantitative evaluation scheme to validate the performance of the comprehensive DL model for GBCA dose reduction.Method
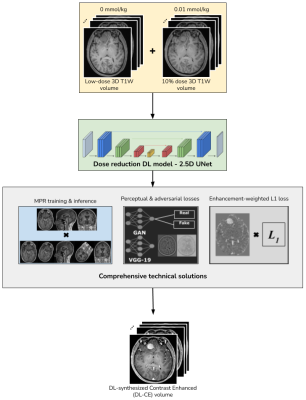
Data & PreprocessingWith IRB approval and patient consent, 3D T1-weighted brain MRI images from 26 patient studies with enhancing tumors were retrospectively selected for this study. These studies were selected from a cohort containing scans from different scanner manufacturers, models and institutional scan protocols. The clinical indications include - meningioma, lymphoma, glioblastoma, etc. As per the scan protocol described in [3], each study contained three 3D T1W scans - pre-contrast, 10% dose (0.01mmol/kg) and full dose contrast-enhanced (CE) (0.1mmol/kg) images. For each patient, a DL synthesized contrast-enhanced (DL-CE) image was obtained using the DL-pipeline in [3], with pre-contrast and 10% dose as inputs. The comprehensive technical solutions applied in the DL model were 2.5D training, multiplanar reconstruction (MPR), perceptual loss, adversarial loss and enhancement weighted loss function, as shown in Fig. 1.
Tumor Segmentation
BRATS[5] is a benchmark dataset containing multimodal images (T1, T1Gd, T2 and FLAIR) of low and high-grade gliomas, with manual annotation of the tumor structures. The goal of the BRATS challenge is to identify the best performing machine learning algorithms that are capable of accurately segmenting the following tumor classes - Whole Tumor (WT), Tumor Core (TC) and Edema/Invasion (ED)[6]. This study uses the top performing model of the BRATS 2018 challenge [7]. This DL model uses a 3D ResNet segmenter with a variational autoencoder (VAE) branch to reconstruct the input image which is used as a regularizer in training the common encoder. The Nvidia GPU Cloud (NGC) [8] has exposed this pre-trained model (trained on the BRATS 2018 data), as a part of their DL model interface. The NGC interface has a variant of this model, using only the T1Gd volume as the input and outputs a binary mask with the Tumor Core (TC) label prediction, which is used in the quantitative evaluation scheme. As shown in Fig. 2, prior to tumor segmentation, the CE and the DL-CE images were skull stripped using the HD-BET tool [9], interpolated to 1mm3 and co-registered to the SRI24 anatomical atlas [10].
Quantitative Evaluation
For each scan, binary masks of TC were derived from both the CE and DL-CE images using the NGC tumor segmentation toolbox. Prior to obtaining the segmentation masks, CE and DL-CE are co-registered to avoid inconsistencies due to slice mismatch. Dice scores were computed between the largest connected components of the segmentation mask of the CE and DL-CE images. The computed Dice scores were used to perform quantitative evaluation. A reasonable Dice score would mean that the dose reduction DL model has the capability of not only reconstructing images with non-inferior image quality, but also without compromising on the diagnostic utility of the synthesized image. Tumor quantification is important for assessing clinical treatment throughout the disease course.
Results
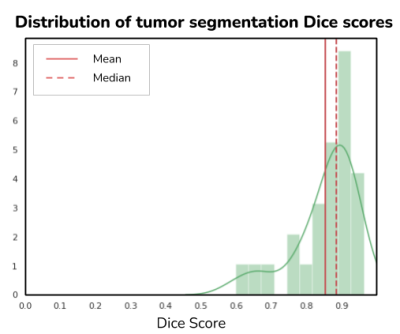
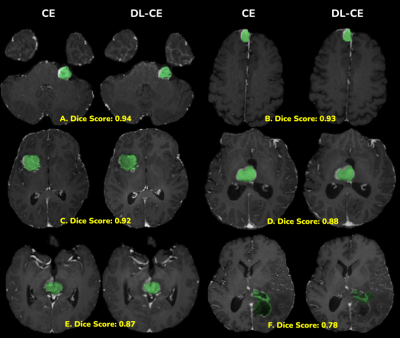
Fig. 3 shows the distribution of Dice scores along with the mean ($0.86 \pm 0.08$) and median 0.89 values. These Dice scores were found to be in the same range as the BRATS 2018 average Dice score of 0.85 for the TC category. The end to end average processing time for a single case was 65 seconds. Fig. 4 shows good visual agreement between the TC masks of CE and DL-CE. Since the BRATS-model was trained only on low & high-grade gliomas, there are some inconsistencies in the TC mask of both CE and DL-CE, even though the enhancement patterns are similar, as shown in Fig. 4F.Discussions
Based on the quantitative scores and visual agreement of the tumor masks, we have demonstrated the feasibility of using a DL-based tumor segmentation method to automate the process of clinical validation of a DL algorithm for dose reduction. Though we have shown examples of enhancing tumors, this scheme can be extended to other clinical indications by using suitable segmentation models.Conclusion
We have proposed an automated quantitative evaluation scheme for the validation of a DL-based contrast dose reduction algorithm.Acknowledgements
We would like to acknowledge the grant support of NIH R44EB027560.References
1) Kanda, T., Ishii, K., Kawaguchi, H., Kitajima, K., Takenaka, D. High Signal Intensity in the Dentate Nucleus and Globus Pallidus on Unenhanced T1-weighted MR Images: Relationship with Increasing Cumulative Dose of a Gadolinium-based Contrast Material. Radiology. 2014; 270(3):834-841.
2) Gong, E., Pauly, J. M., Wintermark, M., Zaharchuk, G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reson Im. 2018; 48(2):330-340.
3) Pasumarthi, S., Tamir, J., Gong, E., Zaharchuk, G., Zhang, T. Toward a site and scanner-generic deep learning model for reduced gadolinium dose in contrast-enhanced brain MRI, Proceedings of the ISMRM 2020. P1286
4) Akkus, Z., Galimzianova, A., Hoogi, A. et al. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J Digit Imaging 30, 449–459 (2017). https://doi.org/10.1007/s10278-017-9983-4
5) Spyridon Bakas, Mauricio Reyes, Andras Jakab, et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv. 2018; preprint:1811.02629.
6) Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K. H. Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. Lect Notes Comput Sc. 2018; 10670:287-297.
7) Andriy Myronenko. 3D MRI brain tumor segmentation using autoencoder regularization. arXiv. 2018; preprint:1810.11654.
8) Nvidia GPU Cloud (NGC). NVIDIA Corporation website. https://www.nvidia.com/en-us/gpu-cloud/, Accessed December 16, 2020.
9) Isensee F, Schell M, Tursunova I, Brugnara G, Bonekamp D, Neuberger U, Wick A, Schlemmer HP, Heiland S, Wick W, Bendszus M, Maier-Hein KH, Kickingereder P. Automated brain extraction of multi-sequence MRI using artificial neural networks. Hum Brain Mapp. 2019; 1–13. https://doi.org/10.1002/hbm.24750.
10) Rohlfing T, Zahr NM, Sullivan EV, Pfefferbaum A. The SRI24 multichannel atlas of normal adult human brain structure. Hum Brain Mapp. 2010 May;31(5):798-819. doi: 10.1002/hbm.20906. PMID: 20017133; PMCID: PMC2915788.
Figures