2158
Mapping Infant Anatomical Brain Images Between Birth and Six Months Using Segmented Brain Images1Department of Pediatrics, Marcus Autism Center, Children's Healthcare of Atlanta, Emory University, Atlanta, GA, United States
Synopsis
Image registration is a critical step for robust and accurate atlas-based analyses on brain volumetric changes. This is especially a challenge in brain growth studies in infants between birth and 6 months of life, when both T1w and T2w magnetic resonance images experience time-varying contrast changes. In this study, we compared image registration outcomes via T1w and tissue segmented images derived using a deep learning method. Our results demonstrated superior registration accuracy based on segmentation images, probably due to clear boundaries between tissues types, as well as their time-constant contrasts over development.
Introduction
Atlas-based volumetric analysis of developing brains using magnetic resonance imaging (MRI) has revealed critical insights into spatial and temporal patterns during early brain development(Gilmore et al., 2007; Knickmeyer et al., 2008). However, during the first six months of life, the gray matter (GM) and white matter (WM) in infant T1- and T2-weighted images experience significant changes in shape and contrast (i.e., iso-intense phase)(Ballesteros et al., 1993). For example, in T1-weighted images, infants at birth show lower signal intensity in unmyelinated WM than in GM, but this GM/WM contrast is somewhat reversed in infants at 6 months or older. Such dramatic changes in both shape and contrasts may impede proper minimization of cost functions during registrations, leading to inaccurate alignment of brain features across subjects or time. Thus, the development of novel strategies that can improve inter-subject registration accuracy during this critical developmental period is urgently needed. In this work, we compared two different approaches for aligning T1 weighted brain images from a group of infants between birth and 6 months. Our results demonstrated that registration based on segmented images of the infant brain in first six months of life may result in more accurate alignment of brain features, when compared to that based on native T1 weighted images.Method
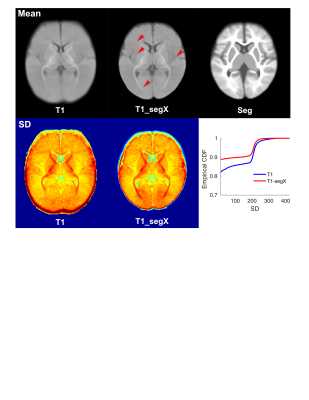
A Siemens 3T Prisma scanner was used to collect T1w and T2w images from 112 typically developing infants between birth and 6 months of age. The MRI protocols for anatomical images were: A 3D T2-weighted SPACE sequence will be acquired with the following parameters: TR/TE=3200/563ms, bandwidth=744 Hz/pixel, echo spacing=3.52ms, Turbo Factor=314, FOV=256×240×167 mm , sagittally acquired, 0.8mm isotropic resolution. The total scan time was 5:57 minutes. T1-weighted images were acquired using a T1-weighted 3D MPRAGE sequence with the following parameters: TR/TE=2400/2.22ms, flip angle=8 degree, FOV=256x240x167mm, 0.8mm isotropic resolution, bandwidth=220 Hz/pixel. Total scan duration was 6:38 minutes. Image segmentation: Deep learning-based methods were applied on combined T1w and T2w images to segment brain images into three tissue types: corticospinal fluid (CSF), gray matter (GM) and white matter (WM)(Wang et al., 2014). Images were registered to a study-specific template using both T1 and segmented images via linear registration tool FLIRT, followed by nonlinear registration tool FNIRT(https://fsl.fmrib.ox.ac.uk/fsl/fslwiki). The detailed steps for generating the study-specific templates using a multi-level, multi-resolution approach are described in(Li et al., 2010). Then, T1 images registered to the original infant space were transformed to the study-specific common space using transformations derived from: 1) T1-weighted (T1) or 2) segmented images (T1_segX). Anatomical details on averaged maps from the group were visually compared. Empirical cumulative density function (ECDF) of the standard deviation map from the two types of templates were also plotted to quantify the registration accuracy.Results
Using the multi-level, multi-resolution registration approach, unbiased study-specific templates were generated using both T1 and segmented images (Fig.1, Top left and Top right). Overall tissue contrasts in both templates are low for infants during first six months of life. Details of myelinated WM in thalamic cortical projections are clearly visible from both templates. Compared to the native T1 template (Fig.1 Top left), the T1 template transformed using the segmented images (Fig.1 Top middle) contained more detailed anatomical features of the brain, such as cortical ribbons (Fig.1, Top middle, red arrows), suggesting more accurate overlapping of corresponding brain structures across infants and time. Empirical CDF of the standard deviation (SD) maps from the two templates further confirmed the hypothesis. The overall SD is lower for the T1_segX template, although both CDF used identical T1 images as inputs.Conclusions
Here, we explored a novel strategy to align infant anatomical brain images into a common space for atlas-based study. We demonstrated that segmented infant brain images may serve as a superior imaging modality for aligning cross-subject and longitudinal images of infant brains, possibly due to their consistent contrasts across development and clear brain tissue boundaries. Future studies should include more publicly available registration tools for evaluation, and the impact of registration on downstream growth curve analysis of early brain volumetric changes.Acknowledgements
We sincerely thank the families and babies for their support. This study is funded by R01EB027147, R01MH119251, 2P50MH100029 and 1R01MH118285References
Ballesteros, M. C., Hansen, P. E., & Soila, K. (1993). MR imaging of the developing human brain. Part 2. Postnatal development. Radiographics, 13(3), 611–622. https://doi.org/10.1148/radiographics.13.3.8316668
Gilmore, J. H., Lin, W., Prastawa, M. W., Looney, C. B., Vetsa, Y. S. K., Knickmeyer, R. C., Evans, D. D., Smith, J. K., Hamer, R. M., Lieberman, J. A., & Gerig, G. (2007). Regional Gray Matter Growth, Sexual Dimorphism, and Cerebral Asymmetry in the Neonatal Brain. Journal of Neuroscience, 27(6), 1255–1260. https://doi.org/10.1523/JNEUROSCI.3339-06.2007
Knickmeyer, R. C., Gouttard, S., Kang, C., Evans, D., Wilber, K., Smith, J. K., Hamer, R. M., Lin, W., Gerig, G., & Gilmore, J. H. (2008). A structural MRI study of human brain development from birth to 2 years. The Journal of Neuroscience, 28(47), 12176–12182. https://doi.org/10.1523/jneurosci.3479-08.2008
Li, L., Preuss, T. M., Rilling, J. K., Hopkins, W. D., Glasser, M. F., Kumar, B., Nana, R., Zhang, X., & Hu, X. (2010). Chimpanzee (Pan troglodytes) precentral corticospinal system asymmetry and handedness: A diffusion magnetic resonance imaging study. PLoS ONE. https://doi.org/10.1371/journal.pone.0012886
Wang, L., Shi, F., Gao, Y., Li, G., Gilmore, J. H., Lin, W., & Shen, D. (2014). Integration of sparse multi-modality representation and anatomical constraint for isointense infant brain MR image segmentation [JOUR]. NeuroImage, 89, 152–164.
Figures