2152
Are rapid structural scans viable for automated lesion assessment in MS patients ?1I2FH, Neuroradiology, CHU Montpellier, University of Montpellier, Montpellier, France, 2Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 3Department of Radiology, Lausanne University Hospital and University of Lausanne, Département de radiologie, Hôpital Universitaire de Lausanne et Université de Lausanne (UNIL), Lausanne, Switzerland, 4LTS 5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 5Siemens SAS Healthcare, Saint-Denis, France
Synopsis
Current MR protocol guidelines for multiple sclerosis (MS) diagnosis and follow-up recommend the acquisition of 3D sequences as high isotropic resolution improves lesion conspicuity; however, this prolongs clinical scan protocols. We present a qualitative and quantitative comparison of lesion assessment using a standard 3D FLAIR and an optimized CAIPIRINHA version in conjunction with compressed sensing MPRAGE in 44 MS patients. We compared the automated lesion segmentation results between protocols, with and without additional manual corrections of the lesion masks. Volumes using the optimized protocol highly correlated with the results from the conventional protocol, while reducing the acquisition time by 47%.
Introduction
Multiple sclerosis (MS) is the most common chronic inflammatory disease of the central nervous system [1]. MRI plays a central role in both diagnosis and follow-up [2, 3]. Recent efforts have focused on standardizing and accelerating imaging protocols to reduce variability, improve patient comfort and allow additional acquisitions beneficial for clinical analysis. In parallel, automated segmentation tools have been developed providing a detailed quantitative analysis of the lesion burden in MS patients [4]. Parallel imaging techniques [5-8] like GRAPPA and CAIPIRINHA [9, 10] have been developed alongside with compressed sensing (CS) , based on a sparse sampling and iterative reconstruction, to accelerate image acquisition even further [11]. This work aims to compare lesion segmentation results obtained with a standard FLAIR sequence and the optimized CAIPIRINHA FLAIR, coupled with a 3D-T1 CS-MPRAGE scan using an automated lesion segmentation tool (LeManPV prototype [12, 13]) on MS patients.Methods
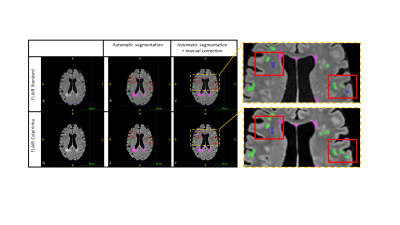
Forty-four MS patients (28 female, age 39 ± 10.5 [18-60]) were included in this monocentric study, all diagnosed by a neurologist (40 relapsing-remitting; 3 primary, 1 secondary progressive; mean EDSS score 1.7 ± 1.9). Examinations were performed at 3T (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) using a 64-channel head coil. Three sequences were acquired for all patients: a GRAPPA 2 accelerated 3D T2 FLAIR (TE/TR/TI: 384/5000/1600 ms, 0.9x0.9x0.9 mm3, acquisition time: 5m22s) and two optimized prototype sequences: 3D-T1 CS-MPRAGE (TR/TI: 2300/900 ms, flip angle 8°, 1x1x1 mm3, acquisition time: 1m43s), and 3D T2 FLAIR CAIPIRINHA with iterative denoising (T2p/TE/TR/TI: 125/415/5000/1650 ms, 1x1x1 mm3, denoising: 85%, acquisition time: 2m51s). Lesion volume was assessed with LeManPV prototype using either reference protocol (3D-FLAIR + 3D-T1 CS) or optimized (3D-FLAIR CAIPIRINHA + 3D-T1 CS) sequences for each patient. Automated segmentation results were manually corrected by an experienced neuroradiologist who was blinded to the imaging protocol (Figure 1). Total lesion volume and count were extracted and compared between conventional and optimized sequences, as well as results with or without manual corrections. We calculated agreement between protocols and segmentation methods using the Lin’s concordance correlation coefficient (CCC) [14]. A CCC greater than 0.99 was considered excellent, 0.95-0.99 substantial, 0.90-0.95 moderate, and less than 0.90 poor. Bland-Altman analyses and Wilcoxon tests were performed to evaluate differences and their significance.Results
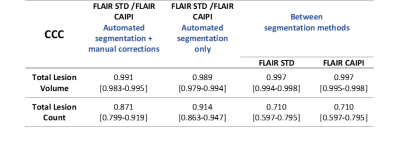
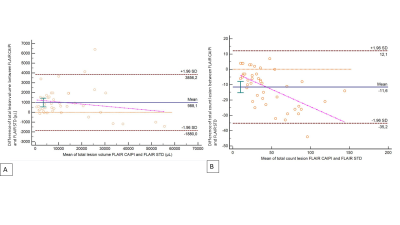
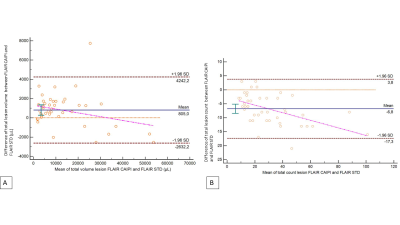
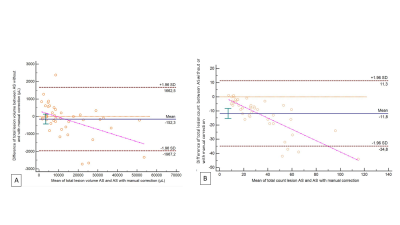
The concordance of total lesion volume between imaging protocols was excellent for both segmentation methods (CCC > 0.99), before and after corrections. Concordance for total lesion count was lower, with a poor concordance between segmentation methods (CCC = 0.71, Table 1). However, we found significant differences for total volume and count between imaging protocols (p<0.01). There was no significant difference for total lesion volume on segmentations before and after manual corrections (p>0.05), but significant for lesion count (p<0.01). Lesion volumes based on FLAIR CAIPIRINHA were systematically higher (Figure 2), with and without corrections (+988 µL [-1880, 3856]; +805 µL [-2632, 4242]), showing larger variations in cases with high lesion load. On the contrary, a lower count of lesions was estimated on FLAIR CAIPIRINHA with and without manual corrections compared to the reference (-11.6 [-35.2, 12.1], -6.8 [-17.3, 3.8], see Figure 3). Total lesion volume and count based on FLAIR CAIPIRINHA were smaller on automated segmentation results compared to results with manual corrections -2.3 µL [-1967.2, 1662.5]/ -11.8 [-34.8, 11.3]) (Figure 4).Discussion
The estimation of total lesion volume presented an excellent concordance, regardless of the FLAIR sequence used or whether manual corrections were applied. Conversely, total lesion counts had poor agreement. This is consistent with previous findings indicating higher variability in lesion count, compared to lesion volume [17]. However, the systematic differences in lesion volumes indicate a different estimation of partial volume (especially in cases with high lesion load) due to increased blurring in the CAIPIRINHA-accelerated sequence after the denoising process. For future work, we would like to investigate the size of missed lesions in relation to the denoising parameters during reconstruction for transversal and longitudinal analyses. Preliminary reviews of the lesions’ masks suggest that the underestimation of lesions differed between brain regions which will be further explored in detail. Moreover, we consider changes at a micrometric scale which may not necessarily have an impact on the clinical assessment. This assumption alongside with the evaluation of the minimum lesion size requires further investigation. Most of the manual corrections were adding small false negatives, especially on the FLAIR CAIPIRINHA sequence. Based on the high agreement on lesion volume between standard and optimized protocols independent of corrections, these preliminary results indicate that automatic lesion segmentation exhibits good overall performance in MS using highly accelerated 3D sequences, where caution should be exercised where small lesions are relevant. Integrating 3D FLAIR CAIPIRINHA and compressed sensing MPRAGE with automated segmentation significantly reduce measurement times for improved patient comfort, less motion susceptibility and more efficient radiological workflow without losing relevant clinical information.Acknowledgements
No acknowledgement found.References
1. Reich DS, Lucchinetti CF, Calabresi PA. Multiple Sclerosis. Longo DL, éditeur. N Engl J Med. 2018;378(2):169-80.
2. Brownlee WJ, Hardy TA, Fazekas F, Miller DH. Diagnosis of multiple sclerosis: progress and challenges. Lancet. 2017;389(10076):1336-1346. doi: 10.1016/S0140-6736(16)30959-X. Epub 2016 Nov 24. PMID: 27889190.
3. Hemond CC, Bakshi R. Magnetic Resonance Imaging in Multiple Sclerosis. Cold Spring Harb Perspect Med. 2018;8(5):a028969.
4. Carass A, Roy S, Jog A, et al. Longitudinal multiple sclerosis lesion segmentation: Resource and challenge. Neuroimage. 2017;148(January):77-102. doi:10.1016/j.neuroimage.2016.12.064
5. Cotton F, Kremer S, Hannoun S, Vukusic S, Dousset V. OFSEP, a nationwide cohort of people with multiple sclerosis: Consensus minimal MRI protocol. J Neuroradiol. 2015;42(3):133-40.
6. Traboulsee A, Simon JH, Stone L, Fisher E, Jones DE, Malhotra A, et al. Revised Recommendations of the Consortium of MS Centers Task Force for a Standardized MRI Protocol and Clinical Guidelines for the Diagnosis and Follow-Up of Multiple Sclerosis. Am J Neuroradiol. 2016;37(3):394-401.
7. Wattjes MP, Rovira À, Miller D, Yousry TA, Sormani MP, de Stefano MP, Tintoré M, Auger C, Tur C, Filippi M, Rocca MA, Fazekas F, Kappos L, Polman C, Frederik Barkhof, Xavier Montalban; MAGNIMS study group. Evidence-based guidelines: MAGNIMS consensus guidelines on the use of MRI in multiple sclerosis--establishing disease prognosis and monitoring patients. Nat Rev Neurol. 2015 Oct;11(10):597-606. doi: 10.1038/nrneurol.2015.157. Epub 2015 Sep 15. PMID: 26369511.
8. Brisset J-C, Kremer S, Hannoun S, Bonneville F, Durand-Dubief F, Tourdias T, et al. New OFSEP recommendations for MRI assessment of multiple sclerosis patients: Special consideration for gadolinium deposition and frequent acquisitions. J Neuroradiol. 2020;47(4):250-8.
9. Breuer FA, Moriguchi H, Seiberlich N, Blaimer M, Jakob PM, Duerk JL, et al. Zigzag sampling for improved parallel imaging. Magn Reson Med. 2008;60(2):474-8.
10. Breuer F, Blaimer M, Griswold M, Jakob P. Controlled Aliasing in Parallel Imaging Results in Higher Acceleration (CAIPIRINHA). Magn Reson Med. 2006;55(3):549-56.
11. Forman C, Wetzl J. Compressed Sensing: a Paradigm Shift in MRI. Magnetom Flash,66, (2016)
12. Fartaria MJ, Bonnier G, Roche A, Kober T, Meuli R, Rotzinger D, et al. Automated detection of white matter and cortical lesions in early stages of multiple sclerosis: Automated MS Lesion Segmentation. J Magn Reson Imaging. 2016;43(6):1445-54.
13. Fartaria MJ, Kober T, Granziera C, Bach Cuadra M. Longitudinal analysis of white matter and cortical lesions in multiple sclerosis. NeuroImage Clin. 2019;23:101938. doi:10.1016/j.nicl.2019.101938 14. Kannengiesser SA, Mailhe B, Nadar M, et al. Universal iterative denoising of complex-valued volumetric MR image data using supplementary information. ISMRM. 2016
15. Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989 Mar;45(1):255-68. PMID: 2720055
16. Paul A. Yushkevich, Joseph Piven, Heather Cody Hazlett, Rachel Gimpel Smith, Sean Ho, James C. Gee, and Guido Gerig. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage 2006 Jul 1;31(3):1116-28.
17. Fartaria, Mário João, Todea, Alexandra, Kober, Tobias, O'Brien, Kieran, Krueger, Gunnar, Meuli, Reto, et al., 2018. Partial volume-aware assessment of multiple sclerosis lesions. NeuroImage 18, 245–253
Figures