2048
Robust estimation of the fetal brain architecture from in-utero diffusion-weighted imaging1Boston Children's Hospital, Harvard Medical School, Boston, MA, United States
Synopsis
Diffusion weighted magnetic resonance imaging (DW-MRI) of fetal brain is challenged by fetal and maternal motion, low signal-to-noise ratio and short scan times. Hence, there is a need for methods that can accurately estimate the parameters of interest from small numbers of noisy measurements. We propose a deep learning method for accurate and robust estimation of color fractional anisotropy as a measure of brain architecture from fetal DW-MRI scans. We also propose methods for simulating realistic training data. Evaluations on an independent cohort of fetal DW-MRI scans show that the proposed method is significantly more accurate than standard methods.
Introduction
Diffusion weighted magnetic resonance imaging (DW-MRI) is routinely used in studying brain microstructure, architecture, connectivity, and abnormalities. There has been growing interest in studying fetal brain development using DW-MRI. In utero DW-MRI, however, is challenged by fetal and maternal motion, limited scan times, limited spatial resolution, and low signal to noise ratio (SNR). Therefore, accurate estimation of parameters of interest from the acquired DW-MRI signal is difficult. Hence, there is an unmet need for methods that can accurately estimate the desired parameters from small numbers of noisy measurements. Recent advancements in deep learning hold great promise for addressing this challenge.The goal of this work was to develop a deep learning method for robust and accurate estimation of color fractional anisotropy (CFA), as a measure of brain architecture, from fetal DW-MRI.
Methods
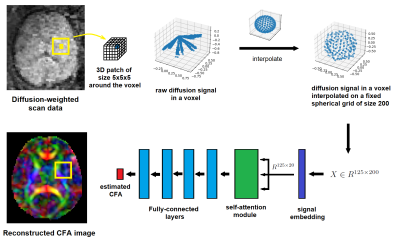
Model:Our proposed model is a deep neural network, shown in Figure 1. In order to exploit the spatial structure of CFA, the model estimates CFA in each voxel based on the measured diffusion signal in a 5x5x5 neighborhood of the voxel. Fetal DW-MRI scans are processed with pipelines that involve motion correction and slice-to-volume registration, resulting in different numbers of diffusion measurements with different gradient directions in each voxel 1. In order for the model to be invariant to these differences and generalizable to different acquisition schemes, the diffusion signal in each voxel is interpolated onto a fixed uniform spherical grid. The network first projects the interpolated signal of each voxel onto a low-dimensional space, where a self-attention module 2,3 learns the complex spatial correlations between signals in neighboring voxels. The output of the attention module goes through a series of fully-connected layers that end in an output layer of size three to predict the three CFA elements.
Training data:
We exploited two auxiliary datasets in generating the training data:
Dataset-1: 36 preterm newborns (gestational age: 29-37 weeks) from the developing human connectome project dataset 4, each with 300 high-quality DW-MRI measurements at three shells: b=0 (n=20), b=400 (n=64), b=1000 (n=88), and b=2600 (n=128). We reconstructed CFA and diffusion tensor images for this dataset using the weighted linear least squares diffusion tensor imaging (WLLS-DTI) method 5.
Dataset-2: 10 fetuses (gestational age: 24-36 weeks) imaged at a single b value of 500 at 24 to 48 gradient directions. We estimated the SNR in fetal DW-MRI using this dataset; the estimated SNR ranged between 14 and 25 dB. Furthermore, we randomly selected 5000 voxels from this dataset and stored their gradient tables. The gradient directions corresponding to each measurement at every voxel was adjusted based on the estimated motion parameters for every slice.
The combination of these two auxiliary datasets enabled us to synthesize realistic training data. CFA images reconstructed with high-quality scans in Dataset-1 enabled us to synthesize data that followed correct spatial neighborhood patterns of CFA. On the other hand, Dataset-2 enabled us to simulate data that followed the SNR and acquisition schemes that matched the diffusion data processed by complicated fetal DW-MRI processing pipelines1.
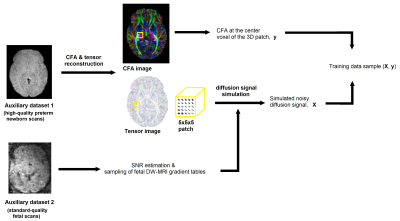
As shown in Figure 2, to simulate a data sample, we selected a 5x5x5 block from a random location in an image from Dataset-1. We used the estimated DTI parameters for the voxels in that block to simulate the diffusion signal using a forward DTI model. The signal was simulated on a fetal gradient table from Dataset-2. Finally, Rician noise with an SNR in the range [14,25]dB was added to the signal for each voxel. We simulated data for five million voxels and used them to train our model.
Training procedures:
The model was trained by minimizing the squared error between the predicted and reference CFA using error back-propagation with Adam optimizer6, batch size of 1000, and learning rate of 0.001 for 100 epochs.
Evaluation:
We tested the trained model on an independent set of 12 fetal scans (gestational age: 23.7-37.1) scanned at a single b-value of 500 at 24 to 60 gradient directions. The scans were processed via motion correction and slice-to-volume registration. We used the complete DW-MRI measurements on these scans to estimate a reference CFA image using WLLS-DTI. We then down-sampled the measurements by discarding 80% of the measurements for each subject. We used our model and WLLS-DTI to reconstruct the CFA images from these down-sampled data and compared the reconstructed images with the reference.
Results
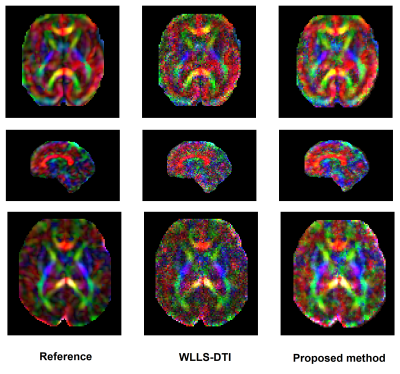
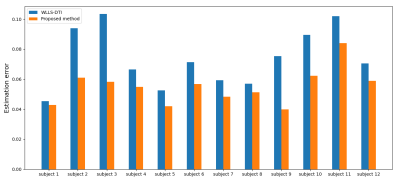
Figure 3 shows example CFA images reconstructed with our method and reference DTI fitting from 80%-down-sampled data, alongside the reference CFA image estimated from full data. Figure 4 shows the root-mean-square of the estimation error for our deep learning method and WLLS-DTI. Our method achieved lower errors on all 12 images. Paired t-test showed that our method was significantly more accurate than WLLS-DTI (p=0.00044).Conclusions
Deep learning methods can significantly improve the parameter estimation accuracy in fetal DW-MRI. In the absence of high-quality real DW-MRI fetal data with ground truth, large amounts of realistic simulated data can be synthesized by exploiting newborn preterm and fetal data.Acknowledgements
This study was supported in part by the National Institute of Biomedical Imaging and Bioengineering, and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (NIH) under Award Numbers R01 EB018988 and R01 NS106030, and a Technological Innovations in Neuroscience Award from the McKnight Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the McKnight Foundation.References
1. Marami, B., Salehi, S.S.M., Afacan, O., Scherrer, B., Rollins, C.K., Yang, E., Estroff, J.A., Warfield, S.K. and Gholipour, A., 2017. Temporal slice registration and robust diffusion-tensor reconstruction for improved fetal brain structural connectivity analysis. NeuroImage, 156, pp.475-488.
2. Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., ... and Polosukhin, I. (2017). Attention is all you need. Advances in neural information processing systems, 30, 5998-6008.
3. Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., ... and Uszkoreit, J. (2020). An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929.
4. Bastiani, M., Andersson, J.L., Cordero-Grande, L., Murgasova, M., Hutter, J., Price, A.N., Makropoulos, A., Fitzgibbon, S.P., Hughes, E., Rueckert, D. and Victor, S., 2019. Automated processing pipeline for neonatal diffusion MRI in the developing Human Connectome Project. NeuroImage, 185, pp.750-763.
5. Koay, C.G., Chang, L.C., Carew, J.D., Pierpaoli, C. and Basser, P.J., 2006. A unifying theoretical and algorithmic framework for least squares methods of estimation in diffusion tensor imaging. Journal of magnetic resonance, 182(1), pp.115-125.
6. Kingma, D. P., Ba, J. (2014). Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.
Figures