2024
Probabilistic structural atlas and connectome of brainstem nuclei involved in arousal and sleep by 7 Tesla MRI in living humans1Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Boston, MA, United States, Charlestown, MA, United States
Synopsis
Brainstem nuclei such as the medullary reticular formation, raphe nuclei, pontine nuclei, locus coeruleus and subcoeruleus, among others, play a crucial role in arousal and sleep. Despite their involvement in critical functions, their study in living humans is challenging due to limited resolution and contrast of conventional MRI. First, we precisely delineated nine arousal/sleep brainstem nuclei in 7 Tesla MRI of healthy humans and generated their validated in-vivo stereotaxic probabilistic atlas. Second, we evaluated their structural connectivity using 7 Tesla High-Angular-Resolution-Diffusion-Imaging and probabilistic tractography. This atlas and connectome might help evaluating disorders of consciousness, sleep disorders and neurological disorders.
Introduction
Several brainstem nuclei are involved in the maintenance of arousal and generation of sleep cycles1. These are the: raphe obscurus (ROb) and pallidus (RPa), involved in neuromodulation2; inferior and superior medullary reticular formation (iMRt, sMRt respectively), involved in arousal and muscle atonia during REM sleep; parvocellular reticular nucleus–Alpha-part (PCRtA) involved in cardiovascular and visceral homeostasis2; subcoeruleus (SubC) generating the REM sleep atonia2; locus coeruleus (LC) involved in arousal, autonomic function, motivation and attention1; pontine reticular formation nuclei (oral and caudal part, PnO-PnC) involved in eye and head movements2, and in the generation and maintenance of REM sleep3; laterodorsal tegmental nucleus (LDTg) involved in neuromodulation and limbic function2. Despite their involvement in critical functions, their study in living humans remains sparse due to the lack of an in-vivo stereotaxic probabilistic atlas of these nuclei.Purpose
Aim1: To create in living humans a stereotaxic probabilistic structural atlas of nine arousal/sleep brainstem nuclei by using high-resolution multi-contrast (diffusion fractional anisotropy —FA— and T2-weighted—T2w) MRI at 7 Tesla. Aim2: To investigate their structural connectivity in living humans by the use of high-spatial and high-angular-resolution-diffusion-imaging (HARDI) at 7 Tesla.Methods
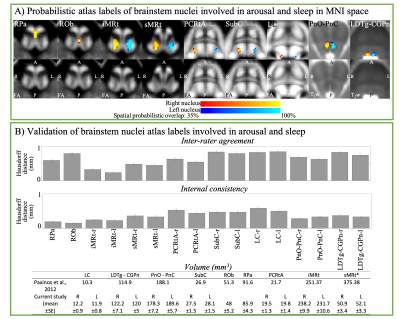
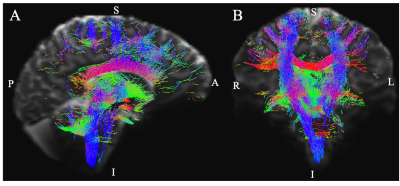
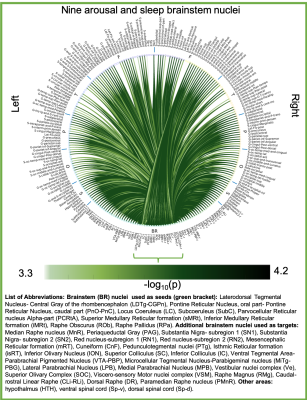
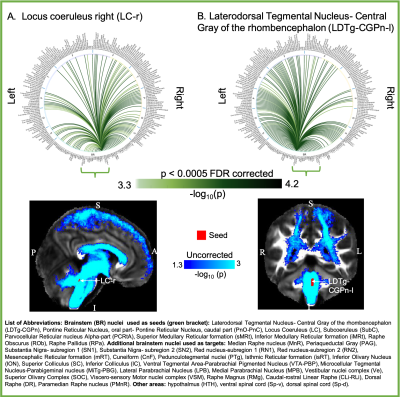
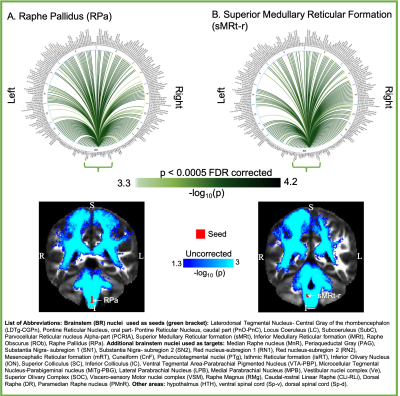
Aim1, data acquisition: Twelve subjects (6m/6f, age 28±1) underwent 7 Tesla MRI under IRB approval. We adopted a common single-shot 2D-EPI readout for 1.1 mm isotropic diffusion-tensor imaging (DTI, using a prototype sequence which supports unipolar diffusion encoding), and T2w sagittal images, with matrix size/GRAPPA-factor/nominal echo-spacing = 180 × 240/3/0.82 ms. This yielded multi-contrast anatomical images with exactly matched resolution and geometric distortions4. Additional MRI parameters were: slices/echo-time/repetition-time/partial-Fourier/n. diffusion-directions/b-value: 61 /60.8 ms/5.6 s/“6/8”/60/ ~ 1000 s/mm2, 7 interspersed “b0” images (non-diffusion weighted, b-value ~ 0 s/mm2), 4 repetitions, acquisition time/repetition 6’43”. Aim1, data analysis: DTIs were rotated to standard orientation, motion- and distortion-corrected (FSL). We then computed the diffusion tensor invariants (e.g. fractional anisotropy, FA) and S0 image (employed as T2w MRI) from the preprocessed DTI (FSL, dtifit). Single-subject FA were coregistered to Illinois-Institute-of-Technology Montreal-Neurological-Institute (IIT-MNI) space through high dimensional non-linear transformations (ANTs)5. On a single-subject basis, two independent raters performed manual segmentation of nine brainstem nuclei on multi-contrast (FA, T2w MRI) images (Figure 1). Voxels rated by both raters as belonging to a nucleus were included in the final nucleus label. A probabilistic atlas for these nuclei in MNI space was created by computing the overlap of nuclei labels across subjects (highest probability = 100 %). Atlas validation: The probabilistic nuclei atlas was validated by the: (i) inter-rater agreement, as the modified Hausdorff distance6 between labels delineated by two raters; (ii) internal consistency across subjects of the final label, as the modified Hausdorff distance between each final label and the probabilistic atlas label (thresholded at 35%) generated by averaging the labels across the other 11 subjects (leave-one-out cross validation); (iii) computation of the nucleus label volume in native space and comparison to literature postmortem volumes7.Aim 2, data acquisition: Nineteen healthy subjects (10m/9f, age 29±5) underwent MRI under IRB-approval using a 7 Tesla scanner. HARDI: common single-shot 2D spin-echo-EPI (using a prototype sequence which supports unipolar diffusion-encoding) with parameters n. slices/echo-time/repetition-time/phase-encoding direction/bandwidth/partial-Fourier/n. diffusion-directions/b-value: 82 /66.8 ms/7.4 s/“anterior/posterior”/“1456 Hz/pixel”/“6/8”/60/2500 s/mm2, seven interspersed “b0” images (T2-weighted, non-diffusion-weighted, b-value = 0 s/mm2), acquisition-time: 8′53′′. To perform distortion-correction we also acquired seven “b0” images with opposite phase-encoding direction. T1-weighted MEMPRAGE image: parameters: repetition-time/echo-times/inversion time/flip-angle/FOV/bandwidth/GRAPPA-factor/ acquisition-time: 2.51s/1.6, 3.5, 5.3, 7.2 ms/1.5 s/7°/256×256×176 mm3/“651 Hz/pixel”/2/6′34′′. Aim 2, data analysis: MEMPRAGE was rotated to standard-orientation, bias field corrected and parcellated with Freesurfer8. HARDIs were de-noised9, motion and distortion-corrected (FSL, topup/eddy). To map the Freesurfer parcellation to native HARDI-space, we computed an affine boundary-based transformation (FSL, FLIRT-BBR) between the preprocessed MEMPRAGE and single-subject S0. To map the brainstem nuclei atlas to native HARDI-space, we computed the bivariate diffeomorphic transformations (ANTs) between IIT-MNI FA/S0 templates10 and single-subject FA/S0images using an intermediate group-based bivariate optimal-template. We performed probabilistic tractography using MRtrix3-iFOD2 algorithm, (90-degree maximum angle between successive steps, 0.07 cut-off, 1-mm minimum streamline-length, Figure 2). We used as seeds the probabilistic labels of nine brainstem nuclei thresholded at 35%, and as targets the Freesurfer parcellations and the nine nuclei labels plus twenty-four additional brainstem nuclei. We propagated 100,000 streamlines from each seed-mask, and computed a “structural-connectivity-index” (range: [0 1]) for each pair of seed-target masks (= fraction of streamlines propagated from the seed reaching the target mask). Wilcoxon test was performed on the connectivity-index values, and significant values were displayed with a 2D circular diagram11.
Results
The probabilistic neuroimaging structural labels in MNI space of the nine brainstem nuclei and their validation are shown in Figure 1. All the investigated brainstem arousal/sleep nuclei displayed specific structural connectivity to cortical, subcortical areas and among themselves (Figures 3-5).Discussion and Conclusions
Our findings demonstrated the feasibility of delineating on a single-subject basis brainstem nuclei in high-contrast and high-sensitivity 7 Tesla MRI, and of generating a validated in-vivo stereotaxic probabilistic atlas of these structures after precise image coregistration to MNI space. We built an in-vivo human 7 Tesla HARDI-MRI based diagram of structural connectivity pathways of these arousal/sleep brainstem nuclei in living humans, in line with animal studies12. This atlas and connectome might help evaluating disorders of consciousness, sleep disorders and neurological disorders.Acknowledgements
MGH-Claflin-Distinguished-Scholar; Harvard-Mind-Brain-Behavior-Faculty-Award; NIH-NIBIB-K01EB019474; NIH-NIDCD-R21DC015888; NIA-R01AG063982. Dr. Thorsten Feiweier for providing the diffusion sequence used in this study.References
1. Satpute A, Kragel PA, Feldman Barrett L, et al., Deconstructing arousal into wakeful, autonomic and affective varieties. Neurosci Lett. 2019; 6;693:19-28.
2. Olszewski J, Baxter D, Karger S. (Ed.), 1954. Cytoarchitecture of the Human Brainstem, JB Lippincott Company, Philadelphia and Montreal North America; 1954.
3. Reinoso-Suárez F, De Andres I, Rodrigo-Angulo ML, et al., Location and Anatomical Connections of a Paradoxical Sleep Induction Site in the Cat Ventral Pontine Tegmentum. European Journal of Neuroscience. 1994; 6: 1829-1836.
4. Bianciardi M, Toschi N, Eichner C, et al. In vivo functional connectome of human brainstem nuclei of the ascending arousal, autonomic, and motor systems by high spatial resolution 7-Tesla fMRI. MAGMA. 2016;29(3):451-62.
5. Avants BB, Tustison NJ, Song G, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033- 2044.
6. Dubuisson, M.P., Jain, A.K., 1994. A modified Hausdorff distance for object matching. Proc. 12th International Conference on Pattern recognition, Jerusalem, Israel, 1, 566-568.
7. Paxinos G, Xu-Feng H, Sengul G, and Watson C. Organization of Brainstem Nuclei. In: The Human Nervous System. Elsevier. 2012.
8. Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968-980.
9. Manjon JV, Coupé P, Concha L, et al., Diffusion Weighted Image Denoising Using Overcomplete Local PCA. PLoS ONE. 2013; 8(9): e73021.
10. Varentsova A, Zhang S, Arfanakis K., Development of a high angular resolution diffusion imaging human brain template. Neuroimage. 2014; 91: 177-186.
11. Irimia A, Chambers MC, Torgerson CM, et al., Circular representation of human cortical networks for subject and population-level connectomic visualization. Neuroimage. 2012; 60: 1340-1351.
12. Bota M, Dong HW, Swanson LW. Combining collation and annotation efforts toward completion of the rat and mouse connectomes in BAMS. Frontiers in Neuroinformatics 2012; 6:2.
Figures