2012
Data-driven bases optimization for fitting of in vivo MR spectra1Radiology, Children's Hospital Los Angeles/USC, Los Angeles, CA, United States, 2Rudi Schulte Research Institute, Santa Barbara, CA, United States
Synopsis
In vivo MR spectra are commonly analyzed by fitting linear combinations of metabolite basis spectra. In this study we explored whether in vivo spectra themselves can provide information that can be used to further optimize basis spectra. Software was developed that includes the capability to modify basis spectra as part of the fitting procedure. The chemical shifts and J-couplings for myo-inositol were optimized by simultaneous fitting of high-quality spectra.
Introduction
In vivo MR spectra are commonly analyzed by fitting linear combinations of metabolite basis spectra. These basis spectra are either obtained from phantom experiments or by computations that simulate MR experiments [1,2]. Generally, these approaches provide very good estimates for the true metabolite spectra. The goal of this study was to evaluate whether basis spectra can be further improved by simultaneous fitting of a large number of high-quality in vivo spectra with J-couplings (JC) and chemical shifts (CS) being additional fit parameters.Methods
Software was developed that allowed the simultaneous fitting of any number of MR spectra. The complete basis set included 25 metabolites as well as signals for lipids and macromolecules. GAMMA [1] was integrated into the software to allow the optimization of chemical shift and J-coupling constants that are used to generate basis spectra. Other fit parameters were the overall frequency alignments of individual spectra, amplitudes of basis spectra, one linebroadening per spectrum, and the chemical shifts of basis spectra relative to each other. The feasibility of fitting individual CSs and JCs was a priori tested and verified with simulated spectra. With these simulations (not shown in detail) we also determined the approximate number of spectra needed to achieve robust true improvements for CSs and JCs for various metabolites and the computational time requirements. It was decided to focus on the CSs and JCs of only myo-inositol (mI) to keep the total fit time within practical limits. Ten spectra of various pathologies that showed prominent mI signal were analyzed. In addition, two separate sets of 20 high-quality grey matter (GM) MR spectra were evaluated. All spectra were acquired on a clinical 3T scanner (Philips Achieva) with single-voxel PRESS, TR=2sec, TE=35ms. Initial CS and JC for basis spectra were taken from [3].Results
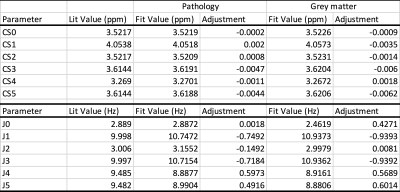
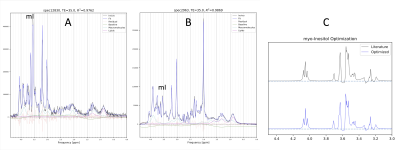
Optimizing CSs and JCs of mI by simultaneous fitting of 10 spectra and 20 spectra took 3h and 16h on a standard PC, respectively. In Table 1 initial CSs and JCs for mI (from [3]) were compared with the results from the simultaneous fitting of 10 pathology and 20 GM spectra. Examples of fitted individual in vivo spectra are shown in Fig. 1A,B whereas the basis spectrum for mI before and after optimization of all CS and JCs is shown in Fig. 1C. Repeating the GM fit with an independent set of 20 spectra gave essentially identical results. Fitting averaged spectra, albeit faster, was less robust.Discussion and Conclusions
Metabolite spectra depend on sequence implementation, acquisition parameters, as well as the microenvironment. Thus, basis spectra that are obtained from model solutions or by computational simulations might be suboptimum. We explored whether in vivo spectra themselves can provide information that can be used to further optimize basis spectra. The initial studies indicate that a slightly modified mI basis spectrum consistently improved the fit across three independent sets of high-quality in vivo spectra. The fitting was highly reproducible when two different sets of grey matter spectra were analyzed. The adjustments for CSs and JCs in abnormal spectra with prominent mI were comparable with observations made for GM spectra. We acknowledge that we cannot rule-out that the modified mI compensates for other imperfections (e.g., missing metabolites in the basis set). Thus, future investigations will include more spectra and the iterative analysis and optimization of other metabolites.Acknowledgements
Rudi Schulte Research InstituteReferences
1. Smith SA., Levante TO, Meier BH, and Ernst RR. (1994). Computer simulations in magnetic resonance. An object- oriented programming approach. J. Magn. Reson. A 106: 75–105
2. Soher BJ, Semanchuk P, Todd D, Steinberg J, Young K. VeSPA: Integrated applications for RF pulse design, spectral simulation and MRS data analysis. Proc. ISMRM 19, 2011.
3. Govindaraju V, Young K, and Maudsley, A.A. (2000). Proton NMR chemical shifts and coupling constants1for brain metabolites. NMR Biomed. 13: 129–153.
Figures