1982
Wave-Encoded Model-Based Deep Learning with Joint Reconstruction and Segmentation1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Havard Medical School, Cambridge, MA, United States, 3National Nanotechnology Center, Pathum Thani, Thailand, 4Harvard/MIT Health Sciences and Technology, Cambridge, MA, United States
Synopsis
We propose a wave-encoded model-based deep learning (wave-MoDL) strategy for simultaneous image reconstruction and segmentation. We use CNN-based regularizers in both k- and image-space to employ features in both domains and successfully incorporated wave-encoding strategy into MoDL reconstruction. Wave-MoDL enables RyxRz=4x4-fold accelerated 3D imaging using a 32-channel array while reducing NRMSE by 1.45-fold compared to wave-CAIPI. Further, we jointly train wave-MoDL and a U-net for simultaneous reconstruction and segmentation to get additional gain.
Introduction
Recently developed model-based deep learning (MoDL) improves image reconstruction by incorporating a convolutional neural network (CNN) into parallel imaging forward model to help denoise and unalias undersampled data(1). Wave-controlled aliasing in parallel imaging (wave-CAIPI) employs extra sinusoidal gradient modulations during the readout to harness coil sensitivity variations in all three-dimensions and achieve higher accelerations(2,3). Wave-encoding strategy was successfully combined with the variational network to provide high-quality images at high acceleration(4,5). In this abstract, we propose wave-encoded (wave-) MoDL and train it jointly with a U-net for simultaneous image reconstruction and segmentation. We incorporated wave trajectory into MoDL reconstruction to better utilize sensitivity encoding. CNNs in both k- and image-space were used to improve the reconstruction by employing features in both domains(6). Further, we take advantage of multi-task learning for joint reconstruction and segmentation, where recent studies reported that training multiple tasks improves the performance of each task(7,8).Methods
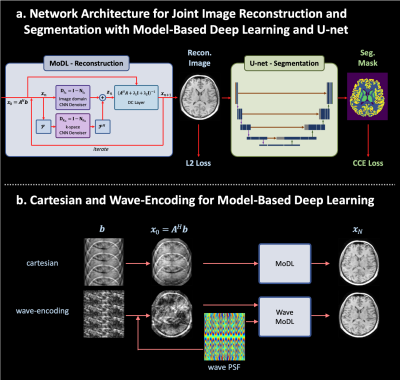
Figure 1a shows the proposed network architecture for joint reconstruction and segmentation with wave-MoDL and U-net. Both mean square error (MSE) and categorical cross entropy (CCE) losses were used to train the network. Figure 1b shows the cartesian- and proposed wave-encodings in the MoDL network. The reconstruction of standard wave-CAIPI is described as follows,$$x = \underset{x}{\operatorname{argmin}}\parallel WF_y PF_x Cx-b\parallel_2^2 $$ $$=\underset{x}{\operatorname{argmin} }\parallel A(x)-b \parallel_2^2 $$
where $$$x$$$ is the reconstructed image, $$$W$$$ is the subsampling mask, $$$P$$$ is the wave point spread function in the kx-y-z hybrid domain, $$$C$$$ is the coil sensitivity map, and $$$b$$$ is the subsampled image, respectively. We used two denoising networks in k- and image-space for wave-MoDL as follows(6).
$$x =\underset{x}{\operatorname{argmin}}\parallel A(x)-b\parallel_2^2 +\lambda_1\parallel N_k(x) \parallel_2^2+ \lambda_2\parallel N_i(x)\parallel_2^2$$
where $$$N_k$$$ and $$$N_i$$$ represents residual CNN networks in the k- and image-space, respectively. We used the alternating minimization-based solution as described in (1,6), by which the network is unrolled as shown in Figure 1a.
To train the network, we used the Calgary-Campinas public dataset(9) and the Human connectome project (HCP) dataset. These datasets include 12-channel GRE images for 67 subjects at 1mm-iso resolution, and 32-channel MPRAGE images for 50 subjects at 0.8mm-iso resolution, respectively. 70%, 15%, and 15% of the subjects in each dataset were used for training, validating, and testing the network, respectively. BART and FREESURFER were used to estimate coil-sensitivity maps and ground-truth segmentation labels(10,11). We aimed to identify four classes, cortical gray matter (GM CTX), subcortical gray matter (GM SUB), white matter (WM), and background (BG). Quadro RTX 5000 was used to train the network. We applied 8-cycle cosine and sine wave-encoding with a maximum gradient strength of 8mT/m, at 200Hz/pixel bandwidth. Example data and code can be found at ‘https://anonymous.4open.science/r/96c9ea60-69b6-4931-8e6a-72a1094626ee’
Results
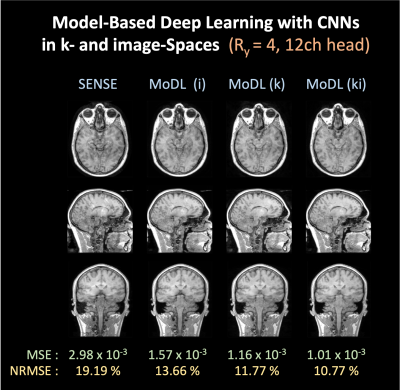
- Model-based Deep Learning reconstruction with CNNs in k- and image-spaceFigure 2 shows MoDL reconstruction at Ry=4 with 12-channel Calgary-Campinas data. MoDL (i), (k), and (ki) represent MoDL with CNN architecture in the image-space, k-space, and both k- and image-space, respectively. For a fair comparison, we used 150K network parameters for all cases; MoDL (i) and (k) have 64 filters in a CNN with 5 layers and MoDL (ki) has 45 filters in each of the two networks with 5 layers. We observed that CNN regularizers in the k-space provide a significant gain for reconstruction. Using two denoisers in the k- and image-space further reduced NRMSE to 10.77%.
- Wave encoded model-based deep learning network (wave-MoDL)
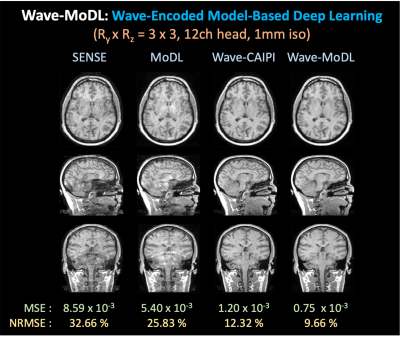
Figure 3 shows the MoDL and wave-MoDL reconstructions at RyxRz=3x3 on 12-channel Calgary-Campinas data. We used two CNN regularizers in the k- and image-space. SENSE failed to reconstruct clean images due to high acceleration in this 12-channel dataset. MoDL improved the images but still suffered from noise amplification and folding artifacts. Wave-CAIPI significantly improved the images by reducing NRMSE of SENSE by 2.65-fold. Wave-MoDL further improved the reconstruction and reduced NRMSE to 9.66%.
- Joint image reconstruction and segmentation
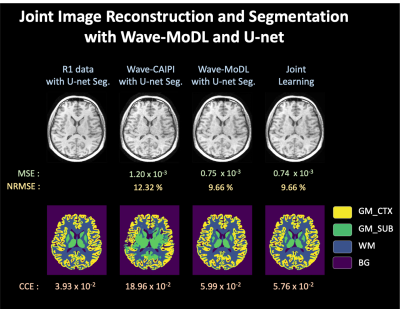
Figure 4 shows joint reconstruction and segmentation with wave-MoDL and U-net on 12-channel Calgary-Campinas data. Using wave-CAIPI images directly as an input to a pre-trained U-net shows poor performance due to noise amplification in the reconstructed images. Combining separately trained wave-MoDL and U-net shows better segmentation results thanks to improved reconstruction. Joint reconstruction and segmentation provided better results on the segmentation task compared with the combination of the separately trained wave-MoDL and U-net.
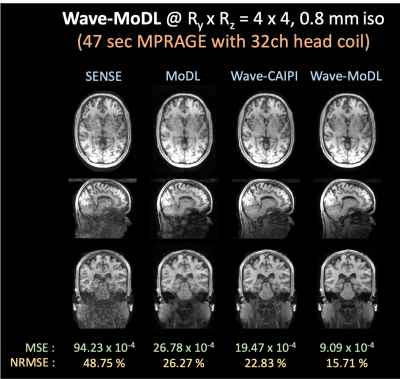
- High accelerated wave-MoDL on 32-channel HCP data
We used wave-MoDL to reconstruct RyxRz=4x4-fold accelerated high-resolution HCP dataset acquired with 32-channel array. SENSE and cartesian-MoDL suffered from noise amplification and artifacts. Wave-CAIPI could significantly improve the results, however lacked SNR at this high under-sampling rate. Wave-MoDL mitigated the noise amplification and reduced NRMSE to 15.71%.
Discussion & Conclusion
We introduced a joint reconstruction and segmentation strategy with wave-MoDL and U-net. Incorporating wave-encoding markedly improved image quality, thus allowing us to push the acceleration to 16-fold on the HCP dataset. Wave-MoDL enabled a 47-second, whole-brain MPRAGE acquisition at 0.8mm-iso resolution with high fidelity.We separated the 3D data into slice groups and trained on sets of aliasing slices. Though this approach is not able to use the information from adjacent slices, it significantly reduces the memory footprint and facilitates training with high channel count data.
Acknowledgements
This work was supported by research grants NIH R01 EB028797, U01 EB025162, P41 EB030006, U01 EB026996 and the NVidia Corporation for computing support. Data collection and sharing for this project was provided by the MGH-USC Human Connectome Project (HCP; Principal Investigators: Bruce Rosen, M.D., Ph.D., Arthur W. Toga, Ph.D., Van J. Weeden, MD). HCP funding was provided by the National Institute of Dental and Craniofacial Research (NIDCR), the National Institute of Mental Health (NIMH), and the National Institute of Neurological Disorders and Stroke (NINDS). HCP data are disseminated by the Laboratory of Neuro Imaging at the University of Southern California.References
1. Aggarwal HK, Mani MP, Jacob M. MoDL: Model-based deep learning architecture for inverse problems. IEEE transactions on medical imaging 2018;38:394–405.
2. Bilgic B, Gagoski BA, Cauley SF, et al. Wave-CAIPI for highly accelerated 3D imaging. Magnetic Resonance in Medicine 2015;73:2152–2162 doi: 10.1002/mrm.25347.
3. Gagoski BA, Bilgic B, Eichner C, et al. RARE/turbo spin echo imaging with simultaneous multislice Wave-CAIPI: RARE/TSE with SMS Wave-CAIPI. Magnetic Resonance in Medicine 2015;73:929–938 doi: 10.1002/mrm.25615.
4. Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data: Learning a Variational Network for Reconstruction of Accelerated MRI Data. Magnetic Resonance in Medicine 2018;79:3055–3071 doi: 10.1002/mrm.26977.
5. Polak D, Cauley S, Bilgic B, et al. Joint multi-contrast variational network reconstruction (jVN) with application to rapid 2D and 3D imaging. Magnetic Resonance in Medicine 2020;84:1456–1469 doi: https://doi.org/10.1002/mrm.28219.
6. Aggarwal HK, Mani MP, Jacob M. MoDL-MUSSELS: Model-Based Deep Learning for Multi-Shot Sensitivity Encoded Diffusion MRI. IEEE Trans. Med. Imaging 2020;39:1268–1277 doi: 10.1109/TMI.2019.2946501.
7. Sun L, Fan Z, Ding X, Huang Y, Paisley J. Joint CS-MRI Reconstruction and Segmentation with a Unified Deep Network. In: Chung ACS, Gee JC, Yushkevich PA, Bao S, editors. Information Processing in Medical Imaging. Cham: Springer International Publishing; 2019. pp. 492–504.
8. Weiss T, Senouf O, Vedula S, Michailovich O, Zibulevsky M, Bronstein A. PILOT: Physics-Informed Learned Optimized Trajectories for Accelerated MRI. arXiv:1909.05773 [physics] 2020.
9. An open, multi-vendor, multi-field-strength brain MR dataset and analysis of publicly available skull stripping methods agreement. NeuroImage 2018;170:482–494 doi: 10.1016/j.neuroimage.2017.08.021.
10. Uecker M, Tamir JI, Ong F, Lustig M. The BART Toolbox for Computational Magnetic Resonance Imaging. :1.
11. Dale AM, Fischl B, Sereno MI. Cortical Surface-Based Analysis: I. Segmentation and Surface Reconstruction. NeuroImage 1999;9:179–194 doi: 10.1006/nimg.1998.0395.
Figures