1978
Training- and Database-free Deep Non-Linear Inversion (DNLINV) for Highly Accelerated Parallel Imaging and Calibrationless PI&CS MR Imaging1Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States, 2UC Berkeley - UC San Francisco Joint Graduate Program in Bioengineering, Berkeley and San Francisco, CA, United States
Synopsis
Deep learning-based MR image reconstruction can provide greater scan time reduction than previously possible. However, this is limited only to MR acquisitions that have large training datasets with fixed hardware and acquisition configurations. We introduce a training- and database-free deep MR image reconstruction technique that may unlock acceleration factors beyond the limits of current state-of-the-art reconstruction methods while being generalizable to any hardware and acquisition configuration. We demonstrate Deep Non-Linear Inversion (DNLINV) on different anatomies and sampling patterns and show high quality reconstructions at higher acceleration factors than previously achievable.
Introduction
Deep learning in image reconstruction has demonstrated state-of-the-art results in MR image reconstruction in the supervised and unsupervised settings. However, a major challenge of deep learning approaches is that there must be a large dataset of raw data to train the models. This limits the access to the benefits of deep image reconstruction to routine protocols with fixed hardware and acquisition configurations. One approach that does not require any training data (and behaves much like classical iterative MR image reconstruction approaches) is the deep image prior (DIP) [1]. The DIP has been demonstrated to provide state-of-the-art results for compressed sensing. This approach is attractive since it does not require any training data and can be applied to any hardware configuration, pulse sequence, or contrast. In this work, we extend the deep image prior approach in two ways: (1) we integrate coil sensitivity estimation in the framework to be applicable to parallel imaging settings, and (2) we apply Bayesian deep learning [2] to make the approach robust to overfitting. We call this approach Deep Non-Linear Inversion (DNLINV).Theory
The DIP approach utilizes a convolutional neural network to generate an image from a set of latent variables that minimizes the mean-square-error with the observed measurements. Based on a U-net architecture, we added an additional branch to the encoded space that estimates coil sensitivity maps as shown in Figure 1. The least-squares DIP objective function is then:$$L = \sum_{i=1}^{N}\sum_{j=1}^K(y_{ij} - PFf_{s_G}(z; \theta)f_{x_G}(z;\theta))^2$$
with the variables defined below, and the optimization is solving for the network parameters $$$\theta$$$. However, this approach is equivalent to maximum likelihood estimation and is sensitive to overfitting amplified noise. We address this by formulating a variational Bayesian inference model to address this limitation. The Bayesian model incorporates noise modeling and the use of the evidence lower bound (ELBO) to control for the complexity of the model and make the model more robust to overfitting. The DNLINV objective function being solved is:
$$
L=-\frac{N}{2}(\log 2\pi +\text{logdet}(\Sigma) - \frac{1}{2\tau}\sum_{i=1}^{\tau}\sum_{j=1}^{K}(y_j - PFf_{s_G}(z; \theta)f_{x_G}(z;\theta))^T \Sigma^{-1} (y_j - PFf_{s_G}(z; \theta)f_{x_G}(z;\theta))- 0.5\sum_{i=1}^{M}\sum_{j=1}^T (z_i^{(j)})^2+ 0.5\sum_{i=1}^{M}\log(\sigma_{z_i}^2) + 0.5
$$
Where N=number of coils, K=number of k-space samples, M=number of latent variables, τ=number of Monte Carlo samples, $$$y_j$$$=row vector of k-space measurements for each coil, P=sampling operator, F=Fourier transform operator, S=Coil sensitivity map operator, Σ=Coil noise covariance matrix, and θ=network parameters. The AdamW [3] optimizer was used to solve for the network parameters and coil noise covariance matrix.
Numerical Experiments
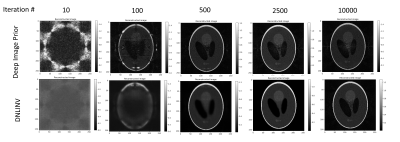
Examining robustness to overfitting: To examine the robustness to overfitting of DNLINV, we simulated a SENSE parallel imaging acquisition of a Shepp-Logan phantom with 8 coils. The data was 3x undersampled along $$$k_y$$$. The image was reconstructed using DIP and with DNLINV using the U-net-like architecture shown in Figure 1. The ground-truth coil sensitivity maps were provided to the reconstruction algorithms to simulate a SENSE reconstruction. The reconstructions at different number of iterations were qualitatively analyzed.Pushing the boundaries of acceleration: We reconstructed two anatomies from the ENLIVE dataset [4]: one knee image acquired with 8 coils and matrix size of 256x320 and one brain image acquired with 32 coils and a matrix size of 192x192. We retrospectively simulated autocalibrated parallel imaging and calibrationless parallel imaging and compressed sensing sampling patterns using the BART toolbox [5] according to the same patterns used in the ENLIVE paper. The sampling patterns are shown alongside the reconstructions in Figures 3 to 5. We reconstructed the PICS data with SAKE [6], ENLIVE [4], and DNLINV, and we reconstructed the autocalibrated parallel imaging data with E-SPIRIT[5], ENLIVE[4], and DNLINV.
Results and Conclusion
Figure 2 shows the results for the SENSE phantom experiment. In the very first few iterations, DIP immediately overfits to the noise due to the noise amplification present in parallel imaging where DNLINV was slowly estimating the image. In later iterations, DIP reconstructs the image however noise amplification artifacts are present whereas DNLINV was able to recover an almost noise-free image and only marginally demonstrates noise amplification artifacts in much later iterations in the optimization.Figure 3 to 4 show the results of DNLINV on calibrationless PICS. At the acceleration factors of 2.0 and 4.0 for the knee and head data, SAKE, ENLIVE, and DNLINV were able to reconstruct the images with high detail. However, at acceleration factors of 5.0 and 8.5, only DNLINV was able to successfully reconstruct the images without any loss of structure. Furthermore, DNLINV produced images with higher apparent SNR compared to the SAKE and ENLIVE.
Figure 5 shows the result for a highly accelerated autocalibrating parallel imaging acquisitions with a CAIPI[7] sampling pattern. At 16x acceleration, ESPIRIT, ENLIVE, and DNLINV were able to successfully reconstruct the image. However, DNLINV demonstrated higher apparent SNR. At 25x acceleration, the ESPIRiT and ENLIVE reconstructions have residual aliasing artifacts whereas these artifacts were largely suppressed in the DNLINV reconstruction.
These results suggest that DNLINV may possibly unlock higher acceleration factors beyond the currently known limits of previous approaches. The DNLINV approach requires no training databases and can be applied to any hardware configuration, pulse sequence, contrast, or sampling scheme.
Acknowledgements
No acknowledgement found.References
[1] D. Ulyanov, A. Vedaldi, and V. Lempitsky, “Deep Image Prior,” ArXiv171110925 Cs Stat, Nov. 2017, Accessed: Jul. 12, 2019. [Online]. Available: http://arxiv.org/abs/1711.10925.
[2] C. Zhang, J. Butepage, H. Kjellstrom, and S. Mandt, “Advances in Variational Inference,” ArXiv171105597 Cs Stat, Oct. 2018, Accessed: Mar. 29, 2020. [Online]. Available: http://arxiv.org/abs/1711.05597.
[3] I. Loshchilov and F. Hutter, “Decoupled Weight Decay Regularization,” presented at the International Conference on Learning Representations, Sep. 2018, Accessed: Dec. 16, 2020. [Online]. Available: https://openreview.net/forum?id=Bkg6RiCqY7.
[4] H. C. M. Holme, S. Rosenzweig, F. Ong, R. N. Wilke, M. Lustig, and M. Uecker, “ENLIVE: An Efficient Nonlinear Method for Calibrationless and Robust Parallel Imaging,” Sci. Rep., vol. 9, no. 1, Art. no. 1, Feb. 2019, doi: 10.1038/s41598-019-39888-7.
[5] M. Uecker et al., “ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA,” Magn. Reson. Med., vol. 71, no. 3, pp. 990–1001, 2014, doi: 10.1002/mrm.24751.
[6] P. J. Shin et al., “Calibrationless parallel imaging reconstruction based on structured low-rank matrix completion: Calibrationless Parallel Imaging,” Magn. Reson. Med., vol. 72, no. 4, pp. 959–970, Oct. 2014, doi: 10.1002/mrm.24997.
[7] F. A. Breuer et al., “Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA),” Magn. Reson. Med., vol. 55, no. 3, pp. 549–556, 2006, doi: 10.1002/mrm.20787.
Figures