1967
PIC-GAN: A Parallel Imaging Coupled Generative Adversarial Network for Accelerated Multi-Channel MRI Reconstruction
Jun Lyu1, Chengyan Wang2, and Guang Yang3,4
1School of Computer and Control Engineering, Yantai University, Yantai, China, 2Human Phenome Institute, Fudan University, Shanghai, China, 3Cardiovascular Research Centre, Royal Brompton Hospital, London, United Kingdom, 4National Heart and Lung Institute, Imperial College London, London, United Kingdom
1School of Computer and Control Engineering, Yantai University, Yantai, China, 2Human Phenome Institute, Fudan University, Shanghai, China, 3Cardiovascular Research Centre, Royal Brompton Hospital, London, United Kingdom, 4National Heart and Lung Institute, Imperial College London, London, United Kingdom
Synopsis
To demonstrate the feasibility of combining parallel imaging (PI) with the generative adversarial network (GAN) for accelerated multi-channel MRI reconstruction. In our proposed PIC-GAN framework, we used a progressive refinement method in both frequency and image domains, which can not only help to stabilize the optimization of the network, but also make full use of the complementarity of the two domains. More specifically, the loss function in the image domain ensures to reduce aliasing artifacts between the reconstructed images and their corresponding ground truth. This enables the model to ensure high-fidelity reconstructions can be obtained even at high acceleration factors.
Purpose
Nowadays, deep learning based models have been exploited to address MRI reconstruction. However, most of the deep learning based studies are limited to single-channel coil raw data. These scenarios are less realistic for clinical applications because modern MRI scanners are normally equipped with multi-channel coils since multi-channel data can provide more imaging information. Thus, this problem remains to be addressed.Introduction
Several endeavors have been made to extend the previous single-channel CNN-based MRI reconstruction methods to the multi-channel reconstruction. Hammernik et al. [1] presented a variational network (VN) for multi-channel MRI reconstruction, and embedded it in a gradient descent scheme. Subsequently, Zhou et al. [2] developed a PI-CNN combined reconstruction framework, which utilized a cascaded structure that intercalated the CNN and PI-DC layer. This method allowed the network to make better use of information from multi-channel coils. Nevertheless, the multi-channel loss function was not integrated into the architecture of the network. Wang et al. [3] trained a deep complex CNN that yielded the direct mapping between aliased multi-channel images and fully-sampled multi-channel images. Unlike other networks for PI, no prior information (such as sparse transform or coil sensitivity) was required, and therefore could provide an end-to-end network for this deep complex CNN based framework. It is of note that all these previous studies have focused only on the single-domain, operating in either the image or the k-space domain, but have not considered both domains simultaneously. In this study, we aim to introduce a novel reconstruction framework named 'Parallel Imaging Coupled Generative Adversarial Network (PIC-GAN)', which is developed to learn a unified framework for improving multi-channel MRI reconstruction.Model Architecture
The schema of the proposed PIC-GAN for multi-channel image reconstruction is illustrated in Figure 1. A deep residual U-Net is adopted for the generator to improve learning robustness and accuracy. A discriminator is connected to the generator output. The discriminator D network is composed of similar encoding part of the generator G.Methods
The proposed architecture, namely PIC-GAN, has integrated data fidelity term and regularization term into the generator so that to fully benefit from information acquired at multi-channel coils and provide an “end-to-end” approach to reconstruct aliasing-free images. Besides, to better preserve image details during the reconstruction, we have combined the adversarial loss with pixel-wise loss in both image and frequency domains. This loss consists of three part, one is a pixel-wise image domain mean absolute error (MAE) $$$L_{i\mathrm{MAE}}(\theta_\mathrm{G})$$$, the other two are frequency domain MAE losses $$$L_{i\mathrm{MAE,R}}(\theta_ \mathrm{G})$$$ and $$$L_{f\mathrm{MAE,1-R}}(\theta_\mathrm{G})$$$. The three loss functions can be written as$$L_{i\mathrm{MAE}}(\theta_\mathrm{G})=\sum_{q}{\left \|x^{q}-\mathrm{S}^{q}\hat{x}_u\right \|_{1}},$$
$$L_{f\mathrm{MAE,R}}(\theta_\mathrm{G})=\sum_{q}{\left \|y^{q}_ \mathrm{R}-\mathrm{R\Im S}^{q}\hat{x}_u\right \|_{1}},$$
$$L_{f \mathrm {MAE,1-R}}(\theta_\mathrm{G})=\sum_{q}{\left \|y^{q}_ \mathrm{1-R}- \mathrm{(1-R)\Im S}^{q}\hat{x}_u\right \|_{1}}.$$
Together with $$$L_\mathrm{adv}$$$, the complete loss function can be written as:
$$L_{\text {total }}=L_{\mathrm{adv}}\left(\theta_{\mathrm{D}}, \theta_{\mathrm{G}}\right)+\alpha L_{\mathrm{iMAE}}\left(\theta_{\mathrm{G}}\right)+\beta L_{\mathrm{fMAE}, \mathrm{R}}\left(\theta_{\mathrm{G}}\right)+\gamma L_{\mathrm{fMAE}, 1-\mathrm{R}}\left(\theta_{\mathrm{G}}\right).$$
Here, $$$q$$$ denotes the coil element, $$$\alpha$$$, $$$\beta$$$ and $$$\gamma$$$ are the hyper-parameters that control the trade-off between each function.
The proposed PIC-GAN framework has been evaluated on abdominal and knee MRI images using 2, 4 and 6-fold accelerations with different sampling patterns. The performance of the PIC-GAN has been compared to the sparsity-based parallel imaging method (L1-ESPIRiT), the variational network (VN) method, and the conventional GAN approach with single-channel images as input (ZF-GAN).
Result
Figure 2 shows representative images reconstructed from ZF, L1-ESPIRiT, VN, ZF-GAN, and PIC-GAN with sixfold undersampling compared to the ground truth (GT). As illustrated in the 1$$$^{st}$$$ and 3$$$^{rd}$$$ rows, the liver and kidney regions are marked with red boxes. The ZF reconstruction was remarkably blurred. Zoomed in error maps showed that liver vessels almost disappeared in L1-ESPIRiT. Moreover, the VN reconstructed images contained substantial residual artifacts, which can be seen in the error maps. The ZF-GAN results produced unnatural blocky patterns for vessels and appeared blurrier at image edges. Compared to the other methods, PIC-GAN results had the least error and were capable of removing the aliasing artifacts.Figure 3 demonstrates the advantage of the proposed PIC-GAN method using different sampling patterns. The ZF reconstructed images presented with a significant amount of aliasing artifacts. Similarly, there were significant residual artifacts and amplified noise that existed in the results obtained by L1-ESPIRiT. For the reconstruction produced by VN, fine texture details were missing, which might limit the clinical usage. The ZF-GAN images enhanced the spatial homogeneity and the sharpness of the images reconstructed by VN. However, ZF-GAN images contained blurred vessels (green arrows) and blocky patterns (yellow arrows). The PIC-GAN not only suppressed aliasing artifacts but also provided sharper edges and more realistic texture details.
As shown in Figure 4, the proposed PIC-GAN method significantly outperformed the L1-ESPIRiT, VN and ZF-GAN reconstruction with an acceleration factor of 2, 4 and 6 with respect to all metrics (p < 0.01) for abdominal data under regular Cartesian sampling.
Conclusion
In conclusion, by coupling multi-channel information and GAN, our PIC-GAN framework has been successfully evaluated using two MRI datasets. Our proposed PIC-GAN method has not only demonstrated superb efficacy and generalization capacity, but has also outperformed conventional L1-ESPIRiT and state-of-the-art CNN based VN and single-channel based ZF-GAN algorithms on 2--6× acceleration.Acknowledgements
This work was supported in part by the National Natural Science Foundation of China No.61902338, in part by IIAT Hangzhou, in part by the European Research Council Innovative Medicines Initiative on Development of Therapeutics and Diagnostics Combatting Coronavirus Infections Award ’DRAGON: rapiD and secuRe AI imaging based diaGnosis, stratification, fOllow-up, and preparedness for coronavirus paNdemics’ [H2020-JTI-IMI2 101005122], and in part by the AI for Health Imaging Award ‘CHAIMELEON: Accelerating the Lab to Market Transition of AI Tools for Cancer Management’ [H2020-SC1-FA-DTS-2019-1 952172].References
- Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data[J]. Magnetic resonance in medicine, 2018, 79(6): 3055-3071.
- Zhou Z, Han F, Ghodrati V, et al. Parallel imaging and convolutional neural network combined fast MR image reconstruction: Applications in low‐latency accelerated real‐time imaging[J]. Medical Physics, 2019, 46(8): 3399-3413.
- Wang S, Cheng H, Ying L, et al. DeepcomplexMRI: Exploiting deep residual network for fast parallel MR imaging with complex convolution[J]. Magnetic Resonance Imaging, 2020, 68: 136-147.
Figures
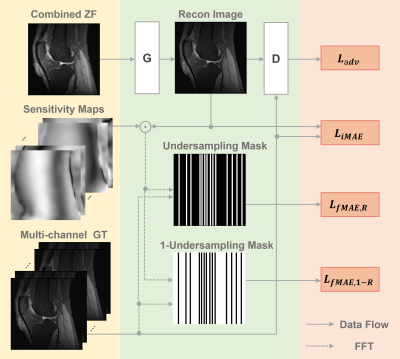
Figure 1. Schema of the proposed parallel imaging and generative adversarial network (PIC-GAN) reconstruction network.
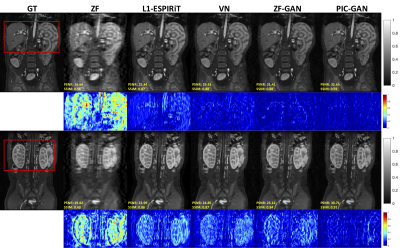
Figure 2. Representative reconstructed abdominal images with acceleration AF= 6. The 1st and 2nd rows depict reconstruction results for regular Cartesian sampling, the 3rd and 4th row depict the same for variable density random sampling. The PIC-GAN reconstruction shows reduced artifacts compared to other methods.
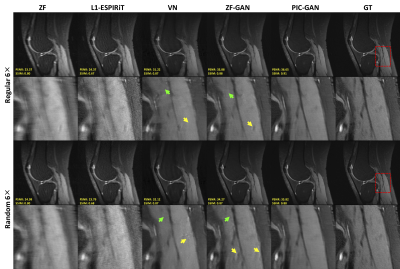
Figure 3. Representative reconstructed knee images with an acceleration factor of 6. The 1st and 2nd rows show reconstruction results using regular Cartesian sampling, the 3rd and 4th rows show reconstruction results using variable density random sampling. Zoomed in views show that the proposed method has resulted in both sharper and cleaner reconstruction compared to the results obtained by L1-ESPIRiT, VN and ZF-GAN. Both ZF-GAN and PIC-GAN reconstruction can significantly suppress artifacts compared to ZF and L1-ESPIRiT.
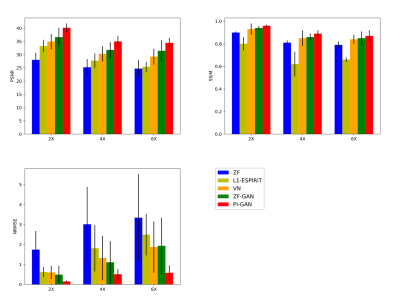
Figure 4. Performance comparisons (NMSE×10-5, SSIM, PSNR and Average Reconstruction Time(s)) on abdominal MRI data with different acceleration factors. The bold numbers highlight the best results. The PIC-GAN outperformed the competing algorithms with significantly higher PSNR, SSIM and lower NMSE values (p<0.05).