1955
Memory-Efficient Learning for High-Dimensional MR Reconstruction1Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 2Pharmaceutical Chemistry, University of California, San Francisco, Berkeley, CA, United States, 3Electrical Engineering, Stanford University, Stanford, CA, United States, 4Radiology, Stanford University, Stanford, CA, United States, 5Electrical and Computer Engineering, The University of Texas at Austin, Austin, TX, United States, 6International Computer Science Institute, University of California, Berkeley, Berkeley, CA, United States
Synopsis
High-dimensional Deep learning (DL) reconstructions (e.g. 3D, 2D+time, 3D+time) can exploit multi-dimensional information and achieve improved results over lower dimensional ones. However, the size of the network and its depth for these large-scale reconstructions are currently limited by GPU memory. Here, we use a memory-efficient learning (MEL) framework, which favorably trades off storage with minimal increased computation and enables deeper high-dimensional DL reconstruction on a single GPU. We demonstrate improved image quality with learned high-dimensional reconstruction enabled by MEL for in-vivo 3D MRI and 2D cardiac cine imaging applications. MEL uses much less GPU memory while minimally increasing training time.
Introduction
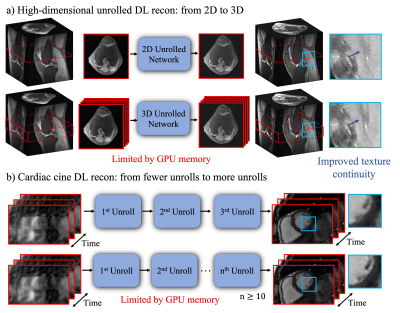
Deep learning-based unrolled reconstructions (Unrolled DL recons)[1-5] have shown great potential for efficient reconstruction from under-sampled k-space data, well beyond the capabilities of parallel imaging and compressed sensing (PICS)[6]. However, so far, implementations have mostly been limited to separable 2D slices. High-dimensional deep learning (DL) reconstructions (e.g. 3D, 2D+time, 3D+time) can take advantage of the multi-dimensional image redundancy in the training data, further improving the image quality. However, these large-scale reconstruction problems are currently limited by the GPU memory required for gradient-based optimization using backpropagation. Therefore, currently, most of the Unrolled DL recons focus on 2D applications or are limited to a small number of unrolls. In this work, we use our recently proposed memory-efficient learning (MEL) framework[7,8] for performing backpropagation, which enables the training of Unrolled DL recons for 1) larger-scale 3D MRI; and 2) 2D+time cardiac cine MRI with a large number of unrolls (Figure 1). We evaluate the spatial-temporal complexity of our proposed method and show that we are able to train high-dimensional MoDL[4] on a single 12GB GPU using much less memory. In-vivo experiments indicate that by exploiting high-dimensional data redundancy, we can achieve improved image quality with sharper edges and better texture continuity for both 3D and 2D+time cardiac cine applications.Methods
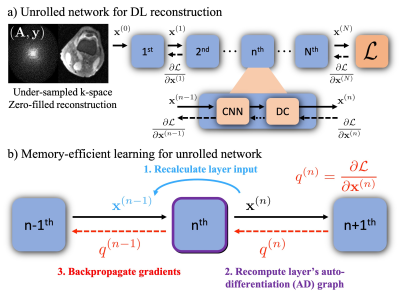
As shown in Figure 2 a), unrolled DL recons often start from under-sampled k-space and zero-filled reconstruction[3,4]. Each unroll consists of two submodules: CNN based regularization layer and data consistency (DC) layer. In conventional backpropagation, the gradient must be computed for the entire computational graph, and intermediate variables from all $$$N$$$ unrolls need to be stored at a significant memory cost. By leveraging MEL, we can process the full graph as a series of smaller sequential graphs. As shown in Figure 2 b), first, we forward propagate the network to get the output $$$\mathbf{x}^{(N)}$$$ without computing the gradients. Then, we rely on the invertibility of each layer (required) to recompute each smaller auto-differentiation (AD) graph from the network’s output in reverse order. MEL only requires a single layer to be stored in memory at a time, which reduces the required memory by a factor of $$$N$$$. Notably, the required additional computation to invert each layer only marginally increases backpropagation runtime.Memory-efficient learning for Model-based Deep Learning (MoDL)
We formulate the reconstruction of $$$\mathbf{x}_{rec}$$$ as an optimization problem and solve it using MoDL:$$\mathbf{x}_{rec}=\arg\min_\mathbf{x}\|\mathbf{Ax-y}\|^2_2+\mu\|\mathbf{x}-R_w(\mathbf{x})\|^2_2,$$
where $$$\mathbf{A}$$$ is the system encoding matrix, $$$\mathbf{y}$$$ denotes the k-space measurements and $$$R_w$$$ is a learned CNN-based denoiser. To apply MEL, $$$R_w$$$ must be invertible (here we used residual CNN[9]). MoDL solves the minimization problem by an alternating procedure:$$\mathbf{z}_n=R_w(\mathbf{x}_n)$$$$\mathbf{x}_{n+1}=\arg\min_\mathbf{x}\|\mathbf{Ax-y}\|^2_2+\mu\|\mathbf{x}-\mathbf{z}_n\|^2_2,$$
which represents the CNN-based regularization layer and DC layer respectively. In order to apply MEL to the above subproblems, the residual CNN-based regularization layer is inverted using a fixed-point algorithm described in [7], while the DC layer is inverted in a closed form as:$$\mathbf{z}_n=\frac{1}{\mu}((\mathbf{A^HA}+\mu\mathbf{I})\mathbf{x}_{n+1}-\mathbf{A^Hy}).$$
Training and evaluation of memory-efficient learning
With IRB approval, we trained and evaluated MEL on both 3D knee and 2D+time cardiac cine reconstructions. We conducted 3D MoDL with and without MEL on 20 fully-sampled 3D knee datasets (320 slices of each) from mridata.org. Around 5000 3D slabs from 16 cases were used for training. Fully-sampled bSSFP cardiac cine datasets were acquired from 15 volunteers at different cardiac views and slice locations. 12 of them (~190 slices) were used for training. We compared the spatial-temporal complexity (GPU memory, training time, inference time) with and without MEL to demonstrate its efficiency. In order to show the benefits of high-dimensional DL recons, we also compared the reconstruction results of 2D and 3D MoDL, 2D+time MoDL with 4 unrolls and 10 unrolls. Peak Signal to Noise Ratio (pSNR) was reported. All the experiments are implemented in Pytorch [10] on Nvidia Titan XP (12GB) and Titan V CEO (32GB) GPUs.
Results
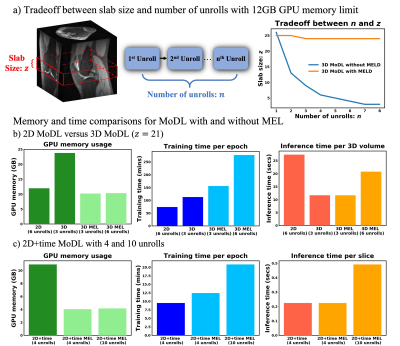
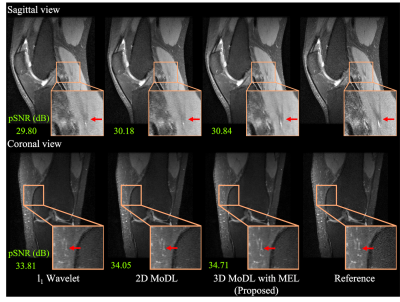
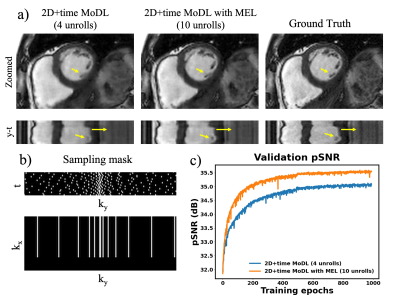
Figure 3 compares the spatial-temporal complexity with and without MEL. Without MEL, for a 12GB GPU memory limit, the maximum slab size decreases rapidly with the increase of unrolls. In contrast, using MEL, the maximum slab size stays roughly the same $$$(\geq24)$$$. Figure 3 b) and c) show that for both 3D and 2D+time MoDL, MEL uses significantly less GPU memory than conventional backpropagation while marginally increasing training time. Figure 4 shows a comparison of different methods for 3D reconstruction. Instead of learning from only 2D axial view slices along the readout dimension (Figure 1 a), 3D MoDL captures the image features from all three dimensions. Zoomed-in details indicate that 3D MoDL with MEL provides more faithful contrast with more continuous and realistic textures. Figure 5 demonstrates that MEL enables the training of 2D+time MoDL with 10 unrolls, which outperforms MoDL with 4 unrolls with respect to image quality and y-t motion profile. Using 10 unrolls over 4 unrolls yields an improvement of 0.6dB in validation pSNR.Conclusions
In this work, we show that MEL enables learning for high-dimensional MR reconstructions on a single GPU, which is not possible with standard backpropagation methods. By leveraging the high-dimensional image redundancy and a large number of unrolls, we were able to recover finer details, sharper edges, and more continuous textures with higher overall image quality.Acknowledgements
We acknowledge support from NIH R01EB009690, NIH R01HL136965, NIH R01EB026136 and GE HealthcareReferences
1. Diamond, S., Sitzmann, V., Heide, F., & Wetzstein, G. (2017). Unrolled optimization with deep priors. arXiv preprint arXiv:1705.08041.
2. Schlemper, J., Caballero, J., Hajnal, J. V., Price, A., & Rueckert, D. (2017, June). A deep cascade of convolutional neural networks for MR image reconstruction. In International Conference on Information Processing in Medical Imaging (pp. 647-658). Springer, Cham.
3. Hammernik, K., Klatzer, T., Kobler, E., Recht, M. P., Sodickson, D. K., Pock, T., & Knoll, F. (2018). Learning a variational network for reconstruction of accelerated MRI data. Magnetic resonance in medicine, 79(6), 3055-3071.
4. Küstner, T., Fuin, N., Hammernik, K., Bustin, A., Qi, H., Hajhosseiny, R., ... & Prieto, C. (2020). CINENet: deep learning-based 3D cardiac CINE MRI reconstruction with multi-coil complex-valued 4D spatio-temporal convolutions. Scientific reports, 10(1), 1-13.
5. Aggarwal, H. K., Mani, M. P., & Jacob, M. (2018). Modl: Model-based deep learning architecture for inverse problems. IEEE transactions on medical imaging, 38(2), 394-405.
6. Lustig, M., Donoho, D., & Pauly, J. M. (2007). Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 58(6), 1182-1195.
7. Kellman, M., Zhang, K., Markley, E., Tamir, J., Bostan, E., Lustig, M., & Waller, L. (2020). Memory-efficient learning for large-scale computational imaging. IEEE Transactions on Computational Imaging, 6, 1403-1414.
8. Zhang, K., Kellman, M., Tamir, I. Jonathan., Lustig, M., & Laura, Waller. (2020, Jan). Memory-efficient learning for unrolled 3D MRI reconstructions. In ISMRM Workshop on Data Sampling & Image Reconstruction, Sedona, AZ.
9. He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778).
10. Paszke, A., Gross, S., Massa, F., Lerer, A., Bradbury, J., Chanan, G., ... & Desmaison, A. (2019). Pytorch: An imperative style, high-performance deep learning library. In Advances in neural information processing systems (pp. 8026-8037).
Figures