1950
Weakly Supervised MR Image Reconstruction using Untrained Neural Networks1Electrical Engineering, Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States
Synopsis
Untrained neural networks such as ConvDecoder have emerged as a compelling MR image reconstruction method. Although ConvDecoder does not require any training data, it requires tens of minutes to reconstruct a single MR slice at inference time, making the method impractical for clinical deployment. In this work, we propose using ConvDecoder to construct "weak labels" from undersampled MR scans at training time. Using limited supervised pairs and constructed weakly supervised pairs, we train an unrolled neural network that gives strong reconstruction performance with fast inference time, significantly improving over supervised and self-training baselines in the low data regime.
Introduction
Supervised MR reconstruction methods such as unrolled neural networks [16,17] currently achieve state-of-the-art image reconstruction performance, often outperforming traditional methods such as parallel imaging [1] and compressed sensing [2]. However, supervised methods are limiting since they require access to training sets with fully-sampled ground truth data, which can be difficult or impossible to acquire.Untrained neural networks have recently shown great promise for image denoising [3-5] and MR image reconstruction [5,6]. These networks do not require any training data; instead, they use the Convolutional Neural Network (CNN) architecture as an image prior for reconstructing natural images. Such networks also provide provable guarantees for denoising and reconstructing smooth signals from few measurements [7,8]. Despite their compelling reconstruction performance that is on-par with some supervised methods, untrained neural networks require tens of minutes to reconstruct a single MR slice at inference time, making them impractical for clinical deployment.
Motivated by the success of weak supervision methods as means for programmatic creation of training sets [9], we propose using ConvDecoder [6], a recently proposed untrained neural network, to generate “weakly-labeled” data from undersampled MR scans at training time. Using few supervised pairs and constructed weakly supervised pairs, we train an unrolled network that gives strong reconstruction performance with fast inference time of few seconds. We show that our method significantly improves over supervised baselines in the low data regime. We also compare against self-training [10-13] (described in Methods), a popular approach that has shown success in computer vision and natural language processing, but has not yet been explored for MR image reconstruction.
Methods
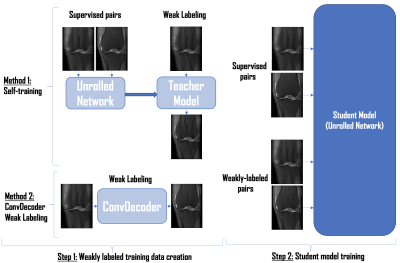
Our proposed two-step weakly supervised MR image reconstruction framework is illustrated in Figure 1. First, we construct weakly supervised pairs from the undersampled scans. Second, we train an unrolled network [18] using limited supervised pairs and the constructed weakly supervised pairs, which is then used at inference time. We compare two methods for creating weakly supervised data: (1) Self-training and (2) ConvDecoder Weak Labeling.(1) Self-training involves training an unrolled neural network using only the supervised pairs, referred to as the “teacher model”. We run inference on undersampled scans using this “teacher model” to create weak labels. These weakly labeled pairs along with supervised pairs are then used to train another unrolled network referred to as the “student model”.
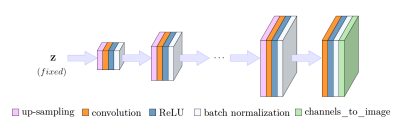
(2) ConvDecoder is an untrained CNN that maps a parameter space to image space $$$G: \mathbb{R}^{p} \rightarrow \mathbb{R}^{c \times w \times h}$$$, as illustrated in Figure 2, where c is number of channels, w and h are width and height of the output image. Input to network is randomly drawn from Gaussian distribution and fixed. Parameters are optimized by fitting the network to a single undersampled measurement according to Equation below, where F is Fourier transform, M is the undersampling mask, $$$S_i$$$ is the coil sensitivity map estimated with SENSE [14], $$$y_i$$$ is the undersampled k-space measurement, and $$$n_c$$$ is the number of coils. Real and imaginary parts of the image are reconstructed in two separate channels, and $$$G(z)$$$ corresponds to the final output complex image. We run 10K iterations of ConvDecoder for each slice of each undersampled volume, and enforce data consistency after the final iteration by enforcing the Fourier transform of the output image to coincide with the original k-space measurements. After using ConvDecoder for weak labeling, we train an unrolled network as our “student model” using both supervised and weakly labeled pairs.
$$\mathcal{L}(z) = \frac{1}{2}\sum_{i=1}^{n_c} \|y_i - MFS_iG(z))\|_2^2 $$
We use a subset of fastMRI knee multicoil dataset [15] for training and evaluation (10 volumes), where the undersampling is conducted retrospectively with a variable density mask with 12 samples for the autocalibration signal and 4x downsampling rate. For training, we investigate using 1 and 3 supervised pairs both with a weakly labeled:supervised ratio of 2:1 (2 and 6 weakly labeled pairs, respectively). We experiment with increasing the weakly labeled:supervised ratio to 5:1 while using 1 supervised pair. We compare our methods with “Supervised Baseline” where we train an unrolled network only using supervised volumes. All unrolled networks are trained with 2 residual blocks for 10 iterations, using L1 reconstruction loss.
Results and Discussion
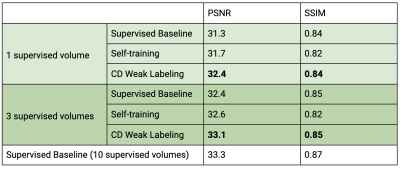
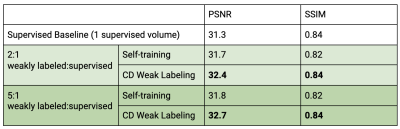
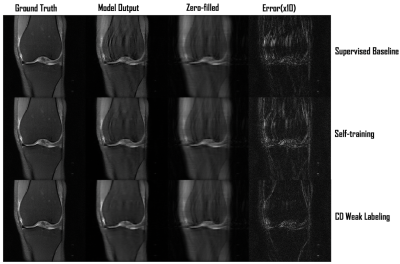
Our framework with ConvDecoder (CD) Weak Labeling improves the Supervised Baseline by 1.1dB and 0.7dB PSNR, and Self-training by 0.7dB and 0.5dB PSNR, when using 1 and 3 supervised volumes, respectively. CD Weak Labeling trained using 3 supervised volumes achieves performance close to supervised training using 10 supervised volumes, as shown in Figure 3. We observe that increasing the weakly labeled:supervised ratio from 2:1 to 5:1 leads to improved performance, resulting in 1.4dB PSNR improvement over the Supervised Baseline using 1 supervised volume, as shown in Figure 4. In Figure 5, we show a comparison for reconstructions where we observe clear improvement in reducing background noise and aliasing artifacts.Our contributions are as follows: (1) We compare self-training, a popular semi-supervised approach, and CD Weak Labeling. (2) Our framework with ConvDecoder Weak Labeling boosts the reconstruction performance, significantly improving over supervised and self-training baselines in the low data regime. It also mitigates the slow inference time for CD while leveraging its image prior biased towards natural images at training time.
Acknowledgements
We would like to acknowledge support from NIH R01EB009690, R01AR07760401, and R01EB026136.References
1. M. Murphy, M. Alley, J. Demmel, K. Keutzer, S. Vasanawala, and M. Lustig. Fast ℓ1-SPIRiT compressed sensing parallel imaging MRI: Scalable parallel implementation and clinically feasible runtime. IEEE Trans Med Imaging. 2012;31(6):1250-1262. doi:10.1109/TMI.2012.2188039
2. M. Lustig, D.Donoho, and JM Pauly. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182-1195. doi:10.1002/mrm.21391
3. D. Ulyanov, A. Vedaldi, and V. Lempitsky. Deep image prior. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pages 9446–9454, 2018.
4. R. Heckel and P. Hand. Deep decoder: Concise image representations from untrained non-convolutional networks. In ICLR 2019.
5. S. Arora, V. Roeloffs, and M. Lustig. Untrained modified deep decoder for joint denoising parallel imaging reconstruction. In ISMRM, 2020.
6. MZ Darestani and R. Heckel. Accelerated MRI with Un-trained Neural Networks. ArXiv abs/2007.02471, 2020.
7. R. Heckel and M. Soltanolkotabi. Denoising and regularization via exploiting the structural bias of convolutional generators. In ICLR 2020.
8. R. Heckel and M. Soltanolkotabi. Compressive sensing with un-trained neural networks: Gradient descent finds the smoothest approximation. In ICML 2020.
9. A. Ratner, SH Bach, HR Ehrenberg, JA Fries, S. Wu, and C. Ré. Snorkel: Rapid Training Data Creation with Weak Supervision. Proceedings of the VLDB Endowment. International Conference on Very Large Data Bases 11 3 (2017): 269-282.
10. B. Zoph, G. Ghiasi, T. Lin, Y. Cui, H. Liu, ED Cubuk, and QV. Le. Rethinking Pre-training and Self-training. In NeurIPS 2020.
11. J. Du, E. Grave, B. Gunel, V. Chaudhary, O. Çelebi, M. Auli, V. Stoyanov, and A. Conneau. Self-training Improves Pre-training for Natural Language Understanding. ArXiv abs/2010.02194, 2020.12. Z. Yalniz, H. Jegou, K. Chen, M. Paluri, and D. Mahajan. Billion-scale semi-supervised learning for image classification. arXiv:1905.00546, 2019.
13. Q. Xie, M. Luong, E. Hovy, and QV. Le. 2020. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 10687–10698.
14. K. P. Pruessmann, M. Weiger, M. B. Scheidegger, and P. Boesiger, “SENSE: Sensitivity encoding for fast MRI,” Magn. Reson. Med., 1999.
15. J. Zbontar et al. FastMRI: An Open Dataset and Benchmarks for Accelerated MRI. ArXiv, abs/1811.08839, 2018.
16. K. Hammernik, T. Klatzer, E. Kobler, MP Recht, DK Sodickson, T. Pock, and F. Knoll. Learning a variational network for reconstruction of accelerated MRI data. Magnetic Resonance in Medicine 2018; 79:3055–3071.
17. HK Aggarwal, MP Mani, and M. Jacob. MoDL: Model-Based Deep Learning Architecture for Inverse Problems. IEEE Transactions on Medical Imaging 2019; 38:394–405.
18. CM Sandino, JY Cheng, F. Chen, M. Mardani, JM Pauly, and SS Vasanawala. Compressed sensing: From research to clinical practice with deep neural networks: Shortening scan times for magnetic resonance imaging. IEEE Signal Processing Magazine, 37(1):117–127, 2020.
Figures