1929
Automatic tract segmentation in the older brain1Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, United Kingdom, 2UCL GOS Institute of Child Health, University College London, London, United Kingdom, 3Department of Psychology, University of Edinburgh, Edinburgh, United Kingdom
Synopsis
TractSeg automatically produces fast and accurate tract segmentations in young populations, but its robustness to changes in the brain due to aging has not been tested. We run TractSeg in data from an older cohort (age 82). We visually assessed the tract segmentations and compared diffusion parameters with a method previously tested in older brains (PNT). TractSeg produced reasonable tract segmentations. The agreement with PNT was poor but measurements were highly correlated. Visual assessments and estimated overlap between segmented bundles suggest potential over-segmentation and subsequent loss of specificity of tract diffusion parameters. Optimised training data could improve TractSeg’s results.
INTRODUCTION
Tractography enables the delineation of brain white matter bundles from diffusion-weighted MRI. Many automatic tractography methods, which require minimal manual intervention, are computationally intensive and complicated to set up through a pipeline of segmentations and registrations. TractSeg is a recent approach based on supervised learning. It is faster and easier to set up, and has been shown to produce accurate segmentations of major white matter tracts1, however it has been trained and tested primarily in data from young brains. We evaluated TractSeg in a sample of older brains to assess its robustness to changes in the brain’s anatomy due to normal aging.METHODS
We used data from the Lothian Birth Cohort 19362, comprising 105 community-dwelling participants (57% male, age 82 years). MRI scans at 3T included a multi-shell diffusion MRI protocol with whole-brain volumes acquired at b = 0, 200, 500, 1000, 2000 s mm-2 and 2mm isotropic voxels. A reverse phase encoding EP dataset was used for susceptibility distortion correction. Data was preprocessed using FSL’s pipeline (‘bet2’, ‘topup’ and ‘eddy’)3–5 and data fitted to a two-fibre model with ‘bedpostX’6.We segmented 10 major white matter tracts: corpus callosum genu and splenium, bilateral anterior thalamic radiation (ATR), arcuate (AF), uncinate (UF) and inferior longitudinal fasciculi (ILF). We ran TractSeg using bedpostX outputs (using the default MRtrix8-based pipeline yielded equivalent results), with both the default set of training tracts (HCP) and the training set derived with FSL’s ‘xtract’7. For comparison we also used probabilistic neighbourhood tractography (PNT)9, which can also use bedpostX outputs and has been previously tested in data from healthy older participants10. We compared the tract segmentations visually using group maps of each tract of interest in MNI space, by registering each tract non-linearly and adding the registered segmentation for all participants. As some level of overlap was observed across different bundles, for each brain we calculated the Jaccard index per pair of tracts (only within the same hemisphere or with corpus callosum, e.g. genu vs left ATR), and the number of overlapping pairs independently for each method. We also measured averaged FA and MD in each tract for each method and calculated rank correlations (rho), and agreement (intraclass correlation coefficient; ICC) between methods per tract. To assess whether differences between methods could be driven by the amount of brain loss due to atrophy, we divided the sample into terciles according to the ratio of brain volume to intracranial volume, and repeated the assessments for each subgroup. We also looked at correlations between mean Jaccard index and brain volume ratio.RESULTS
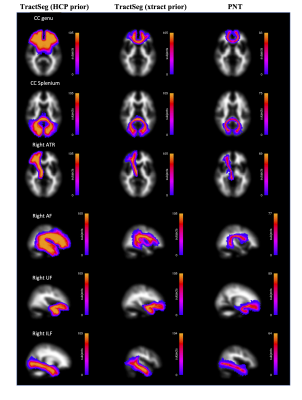
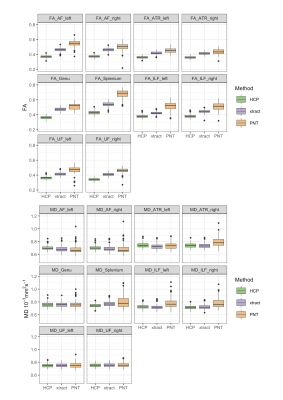
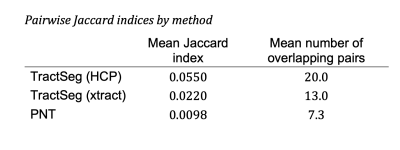
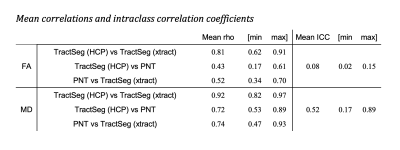
Figure 1 shows the group maps. TractSeg using HCP priors consistently segmented a larger area of white matter, while PNT segmentations were mainly contained within the centre of the bundle. This was reflected in the overlap between tracts, as shown by the mean Jaccard index obtained for each method, and the number of overlapping tract pairs (Table 1). The diffusion parameters measured in each tract for each method are represented in Figure 2. PNT tracts had the highest FA and TractSeg (HCP) the lowest for all tracts. The mean ICC calculated for the three methods was 0.08 (range [0.02, 0.15]), confirming the poor agreement between them. The MD values showed better agreement, with mean ICC of 0.52 ([0.17,0.89]), with the lowest agreement corresponding to the right ATR and the highest to the UF. Nevertheless, the values were highly correlated across methods as shown by the mean rho for each pair of methods in Table 2, with the two TractSeg methods being more correlated. After dividing the sample into brain volume ratio terciles, the group maps, ICCs and correlations calculated within each subgroup did not show any patterns across terciles, suggesting that the difference observed between methods might not be just driven by age-related brain loss. However, we found that the Jaccard index was correlated with brain volume ratios for TractSeg approaches (rho=0.30 for HCP, 0.28 for xtract, both p<0.004), but not for PNT (rho=-0.01,p=0.91).DISCUSSION
TractSeg produced reasonable segmentations in the older brain. Some overlap between tracts may be expected in areas of crossing fibres, but extensive overlap may be indicative of over-segmentation. This reduced the specificity of this method and could explain the lower variability in FA and MD between tracts, tending towards a more global measure of brain microstructural integrity. (Whole brain FA averaged 0.224 in this dataset; MD averaged 1.15×10-3 mm2s-1). Using ‘xtract’ training tracts produced tighter bundles. PNT produced more specific bundles, with greater variability in parameters between tracts, but with the trade-off of not covering their whole extent. Although the modelling approaches are quite different, PNT models were trained in an older dataset (71–73y)10 compared to the HCP dataset (22–35y) used to train both TractSeg and xtract. We did not observe a link between brain loss and agreement between methods, but it was related to the degree of overlap between tracts, so using older brain data for training might reduce over-segmentation in this population.CONCLUSION
There are multiple ways of segmenting white matter tracts, and all of these automated techniques produced plausible results overall. The trade-off for individual studies is between more inclusive segmentations and greater tract specificity.Acknowledgements
The Lothian Birth Cohort 1936 phenotype collection was supported by Research Into Ageing, Age UK, and the UK Medical Research Council. MRI acquisition was conducted at the Edinburgh Imaging Facilities, University of Edinburgh. This work was supported by the Centre for Cognitive Ageing and Cognitive Epidemiology, funded by the Medical Research Council and the Biotechnology and Biological Sciences Research Council the Row Fogo Charitable Trust, the European Union Horizon 2020, PHC-03-15, SVDs@Target, the Fondation Leducq Transatlantic Network of Excellence for the Study of Perivascular Spaces in Small Vessel Disease, and the Medical Research Council UK Dementia Research Institute at the University of Edinburgh. We thank the Lothian Birth Cohort 1936 participants who took part in this study, the research team members, and radiographers at the Edinburgh Imaging Facilities.References
1. Wasserthal, J., Neher, P. and Maier-Hein, K. H. TractSeg - Fast and accurate white matter tract segmentation. Neuroimage 183, 239–253 (2018).
2. Taylor, A. M., Pattie, A. and Deary, I. J. Cohort Profile Update: The Lothian Birth Cohorts of 1921 and 1936. Int. J. Epidemiol. 47, 1042-1042r (2018).
3. Smith, S. M. Fast robust automated brain extraction. Hum. Brain Mapp. 17, 143–155 (2002).
4. Andersson, J. L. R., Skare, S. and Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20, 870–888 (2003).
5. Andersson, J. L. R. and Sotiropoulos, S. N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 125, 1063–1078 (2016).
6. Behrens, T. E. J., Johansen-Berg, H., Jbabdi, S., Rushworth, M. F. S. and Woolrich, M. W. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage 34, 144–155 (2007).
7. Warrington, S., Bryant, K. L., Khrapitchev, A. A., Sallet, J., Charquero-Ballester, M., Douaud, G., Jbabdi, S., Mars, R. B. and Sotiropoulos, S. N. XTRACT - Standardised protocols for automated tractography in the human and macaque brain. Neuroimage 217, 116923 (2020).
8. Tournier, J. D., Smith, R., Raffelt, D., Tabbara, R., Dhollander, T., Pietsch, M., Christiaens, D., Jeurissen, B., Yeh, C. H. and Connelly, A. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage 202, 116137 (2019).
9. Clayden, J. D., King, M. D. and Clark, C. A. Shape Modelling for Tract Selection. in Lecture Notes in Computer Science, vol 5762 (eds. GZ., Y., D., H., D., R., A., N. & C., T.) 150–157 (Springer, Berlin, Heidelberg, 2009).
10. Muñoz Maniega, S., Bastin, M., Deary, I., Wardlaw, J. and Clayden, J. Reference Tracts and Generative Models for Brain White Matter Tractography. J. Imaging 4, 8 (2018).
Figures