1916
Brain rejuvenation improves accuracy of automatic segmentation of the aged brain1Radiology, Albert Einstein College of Medicine, Bronx, NY, United States
Synopsis
Despite best efforts, performance of automatic brain segmentation is inferior in older adults. We hypothesize that worsening of segmentation quality with age is related to brain atrophy and changes to image contrast. To disentangle the two possibilities, we simulated brain atrophy and confirmed worse segmentation performance. We then propose and demonstrate “brain rejuvenation” as a method to improve segmentation of the aged brain.
Introduction
Segmentation of brain regions into a subject specific atlas is a widely used approach to image analysis. FreeSurfer1 is one of the most widely used and validated tools to automate this tedious and difficult process. Despite best efforts, FreeSurfer accuracy declines in older adults2-5. When compared with manual segmentation of the hippocampus, for example, FreeSurfer showed under-estimation of aged brain volumes3. In another study, Freesurfer validly predicted the size of young hippocampi , but underestimated larger hippocampal volumes in older adults4. Manual correction of the Freesurfer segmentation can be applied to improve Freesurfer accuracy, but is time-consuming and cannot be automated2.We hypothesize that worsening of segmentation quality is function of age-related brain atrophy and/or change of image contrast. To disentangle the two, we confirmed worsening of FreeSurfer segmentation performance in the presence of simulated brain atrophy. We then propose and demonstrate “Brain Rejuvenation” as a method to improve segmentation of the aged brains.
Methods
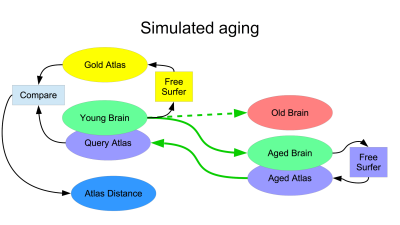
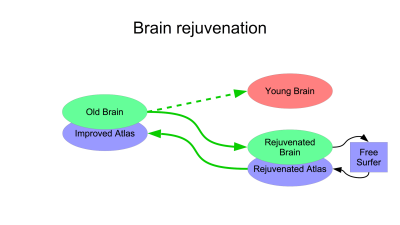
To simulate brain atrophy (Figure 1), we morphed the (young) JHU template brain to a set of images of various ages, segmented the results using the ASEG module of FreeSurfer1 version 6.0, transformed the segmentations back to the JHU template brain and compared this to its native FreeSurfer segmentation as the gold standard. Comparison was based on displacement of anatomical landmarks6 (blue in Fig 1). Brain rejuvenation entails this same process in reverse (Figure 2): an elderly brain is morphed to a young brain creating the rejuvenated brain, which is segmented using FreeSurfer. This segmentation is transformed back to the original aged brain.This study was approved by IRB and includes 165 T1W images obtained as part of ongoing lifespan studies (age range: 18-91, 92 females). These images are used as targets of simulated aging to cover wide range of brain atrophies. All images were acquired on a 3.0T Philips Achieva TX scanner (Philips Medical Systems, Best, The Netherlands) utilizing its 32-channel head coil with the following protocol: TR/TE/TI = 9.9/4.6/900 msec, flip angle 8deg, 1mm3 isotropic resolution, 128x116x220 matrix. All images were reviewed by an experienced neuroradiologist and determined to be free of visible structural abnormalities. ANTs symmetric normalization7 with parameters 3, 0, 0.9 was used to compute morphisms between brains and their inverses.
To illustrate improved segmentation of aged brains using brain rejuvenation, we repeated the procedure shown in Figure 1, replacing segmentation of the aged brain by FreeSurfer (purple FreeSurfer on the right of Figure 1) with the proposed brain rejuvenation of Figure 2.
Results
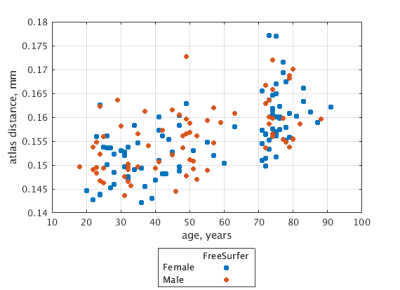
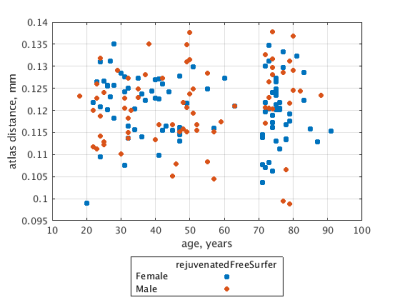
Figure 3 illustrates sensitivity of FreeSurfer segmentation to brain atrophy indexed by the age of the target aged brain; atlas distance, with respect to the gold-standard, increases with the age of the target. This age dependence is rectified (Figure 4) when the proposed brain rejuvenation (Figure 2) is used instead of FreeSurfer.Conclusion.
Consistent with the previously observed worsening of quality of segmentation of aged brains, our results indicate that FreeSurfer accuracy is specifically sensitive to age-related brain atrophy. Our proposed brain rejuvenation rectifies this sensitivity.Acknowledgements
Support for this research was provided in part by the National Institute on Aging, grant 5P01AG003949-34.References
1. Fischl B., Salat D. H., Busa E., et al. Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain. Neuron 2002; 33: 341.
2. Huo Y, Plassard A, Carass A, et al. Consistent cortical reconstruction and multi-atlas brain segmentation. Neuroimage. 2016 Sep;138:197-210.
3. Srinivasan D, Erus G, Doshi J, et al. A comparison of Freesurfer and multi-atlas MUSE for brain anatomy segmentation: Findings about size and age bias, and inter-scanner stability in multi-site aging studies. Neuroimage. 2020 Dec;223:117248.
4. Wenger E, Mårtensson J, Noack H, et al. Comparing manual and automatic segmentation of hippocampal volumes: Reliability and validity issues in younger and older brains. Hum Brain Mapp. 2014 Aug;35(8):4236-48.
5. Waters AB, Mace RA, Sawyer KS, Gansler DA. Identifying errors in Freesurfer automated skull stripping and the incremental utility of manual intervention.Brain imaging and behavior 13(5), 1281-1291 (2019).
6. Gil N, Lipton ML, Fleysher R. Registration Quality Filtering Improves Robustness of Voxel-Wise Analyses to the Choice of Brain Template. NeuroImage 2020, in press.
7.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033-44.
Figures