1867
Automated pancreas sub-segmentation by groupwise registration and minimal annotation enables regional assessment of disease
Alexandre Triay Bagur1,2, Ged Ridgway2, Sir Michael Brady2,3, and Daniel Bulte1
1Department of Engineering Science, The University of Oxford, Oxford, United Kingdom, 2Perspectum Ltd, Oxford, United Kingdom, 3Department of Oncology, The University of Oxford, Oxford, United Kingdom
1Department of Engineering Science, The University of Oxford, Oxford, United Kingdom, 2Perspectum Ltd, Oxford, United Kingdom, 3Department of Oncology, The University of Oxford, Oxford, United Kingdom
Synopsis
A method to automatically segment the pancreas into its main subcomponents head, body and tail is presented. The method uses groupwise registration to a reference template image that is subsequently annotated by parts. A new subject is registered to the template image, where part labels are propagated, and then transformed back to subject space. We test the method on the UK Biobank imaging sub-study, using a nominally healthy all-male cohort of 50 subjects for template creation and 20 other subjects for validation. We show pancreas T1 quantification by segment when reslicing the segmentation on a separate T1 slice.
Introduction
Pancreas disease, including fat infiltration, fibro-inflammation, and pancreatic cancer, is generally heterogeneous. Circulating biomarkers of pancreas disease are non-specific, while invasive tests are harmful and may lead to complications. Unlike MRI, none can give information on disease heterogeneity. Many studies have reported clinically important differences in the head, body and tail parts using regions of interest1,2. We show that we can automatically segment these parts in MRI, further enabling such work. Our method is based on groupwise registration, and requires minimal annotation. We validate the method on the UK Biobank imaging sub-study.Automated segmentation methods, including deep learning-based, have to date aimed at delineating the whole pancreas, and reported excellent accuracy3. Fully automated approaches for partitioning the pancreas into head, body and tail subcomponents have not been reported (though semi-automated methods exist4). Full automation would be particularly useful where manual intervention is too costly or is infeasible, such as in big data studies like UK Biobank5.
Methods
We gathered a ‘training’ dataset comprising N=50 subjects from UK Biobank (Siemens Aera, 1.5 T); all were nominally healthy male subjects aged 50 to 70 (Fig. 1A). We first performed whole-pancreas segmentation on all subjects using a deep learning-based method presented elsewhere6 (and available online on GitHub7) applied on the volumetric interpolated breath-hold examination (VIBE) scan.We then groupwise-registered the segmentations into a group mean (‘template’) using the Large Deformation Diffeomorphic Metric Mapping (LDDMM) via Geodesic Shooting8, available in the “Shoot” toolbox of SPM129. We manually annotated pancreas head, body and tail on the template volume, so that only 1 annotation step was required for the entire dataset. Annotation was performed using the 3D scalpel tool of ITK-SNAP10, defining one separation plane between head and body and one between body and tail (Fig. 1A). Then, for any new subject, the method: (1) segments the whole pancreas; (2) warps the segmentation to the template; (3) propagates the template part labels to the warped subject; and (4) warps the warped subject’s parts segmentation back to subject space using the inverse transformation (Fig. 1B–E).
We gathered a ‘validation’ dataset of N=20 other subjects from UK Biobank with the same demographics as the training dataset, and performed parts segmentation using our method (Fig. 2). We evaluated the partitions using reference manual annotations performed on each subject individually, twice by the same user, so that the intra-observer performance can be used as a comparative upper bound on the automatic results.
Results
Segmentation performance is commonly evaluated with Dice. However, we assume whole-pancreas segmentation is available, and the aim is to find the 2 separation boundaries between the 3 pancreas parts, but not their outer boundary (Fig. 3). Thus, we used the root mean squared error (RMSE) of the Euclidean distance between the 2 point clouds defined by the annotation boundary and the predicted boundary (Fig. 4A). Point correspondence was established using MATLAB’s Iterative Closest Point algorithm (pcregistericp). We also reported Hausdorff distance (HD), recognising that only HD in head and tail are truly independent measurements (Fig. 4B).Intra-observer distance (annotation 1 vs annotation 2) was used as upper threshold performance. For both RMSE and HD metrics, results between intra-observer variability and parts segmentation are comparable. Furthermore, no significant difference was found for the head/body boundary (p=0.710 RMSE, p=0.084 HD). A significant difference between intra-observer annotations and predictions was observed for the body/tail boundary (p=0.037 RMSE, p=0.005 HD), that we discuss below.
Figure 5 shows the feasibility of regional quantification of pancreas imaging biomarkers using the parts segmentation from our method. The three-dimensional pancreas parts segmentation of a subject in UK Biobank was resliced onto the T1 map from a separate breath-hold scan, resulting in a two-dimensional mask with the 3 pancreas segments; segment medians may be reported.
Discussion
We chose to work initially with UK Biobank data comprising nominally healthy volunteers aged 50 to 70, and selected an all-male cohort with no self-reported diabetes of any type, though we plan to expand this cohort in future validation. While the VIBE scan is high-quality, it often has limited coverage of the pancreas head6, though this is consistent across subjects. Annotations from a different expert will be collected in future work for an inter-rater measure of performance for comparison.Our modular approach to parts segmentation facilitates replacement of the upstream whole-organ segmentation when a better method becomes available, reduces problem complexity and expedites validation. RMSE of Euclidean distance was reported, though we may choose to fit each predicted boundary to a plane – like the manual annotations – that is orthogonal to the pancreas medial axis; in this scenario, the distance reduces to a scalar offset. Furthermore, since the body-tail boundary is “generally agreed to be located at the midpoint of the total length of the body and tail”11, we may further break down our task to identifying the head-body boundary, for which we show good results (Fig. 4).
Conclusion
This study demonstrated the feasibility of automated pancreas parts segmentation by using groupwise registration of whole-organ segmentations to a template, and the subsequent annotation of the template image. This enables segmental characterisation of pancreas disease.Acknowledgements
Perspectum Ltd for providing funding, data and counseling, as well as the Engineering and Physical Sciences Research Council (EPSRC) for providing funding. This research has been conducted using the UK Biobank Resource under Application Number 9914.References
- Nadarajah C, Fananapazir G, Cui E, et al. Association of pancreatic fat content with type II diabetes mellitus. Clin Radiol. 2020;75(1):51-56. doi:10.1016/j.crad.2019.05.027
- Tirkes T, Lin C, Fogel EL, Sherman SS, Wang Q, Sandrasegaran K. T1 mapping for diagnosis of mild chronic pancreatitis. J Magn Reson Imaging. 2017;45(4):1171-1176. doi:10.1002/jmri.25428
- Oktay O, Schlemper J, Folgoc L Le, et al. Attention U-Net: Learning Where to Look for the Pancreas. 2018;(Midl):1-10. http://arxiv.org/abs/1804.03999
- Fontana G, Riboldi M, Gianoli C, et al. MRI quantification of pancreas motion as a function of patient setup for particle therapy -A preliminary study. J Appl Clin Med Phys. 2016;17(5):60-75. doi:10.1120/jacmp.v17i5.6236
- Littlejohns TJ, Holliday J, Gibson LM, et al. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nat Commun. 2020;11(1):1-12. doi:10.1038/s41467-020-15948-9
- Bagur AT, Ridgway G, McGonigle J, Brady SM, Bulte D. Pancreas Segmentation-Derived Biomarkers: Volume and Shape Metrics in the UK Biobank Imaging Study. In: Communications in Computer and Information Science. Vol 1248 CCIS. ; 2020:131-142. doi:10.1007/978-3-030-52791-4_11
- Pancreas Segmentation in UK Biobank. Accessed December 16, 2020. https://github.com/alexbagur/pancreas-segmentation-ukbb
- Ashburner J, Friston KJ. Diffeomorphic registration using geodesic shooting and Gauss-Newton optimisation. Neuroimage. 2011;55(3):954-967. doi:10.1016/j.neuroimage.2010.12.049
- SPM12 - Statistical Parametric Mapping. Accessed December 16, 2020. https://www.fil.ion.ucl.ac.uk/spm/software/spm12/
- Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116-1128. doi:10.1016/j.neuroimage.2006.01.015
- Suda K, Nobukawa B, Takase M, Hayashi T. Pancreatic segmentation on an embryological and anatomical basis. J Hepatobiliary Pancreat Surg. 2006;13(2):146-148. doi:10.1007/s00534-005-1039-3
Figures
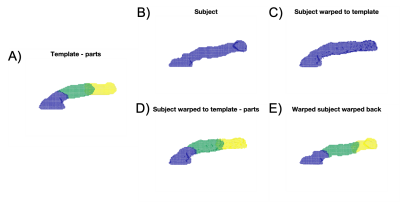
Figure 1. Groupwise registration of the whole-pancreas segmentation generated a group mean (‘template’), on which the parts were manually annotated (A, head: blue, body: green, tail: yellow). For any new subject, the method segments the whole pancreas (B); warps the segmentation to the template (C); propagates the template part labels to the warped subject (D); and warps the warped subject’s parts segmentation back to subject space using the inverse transformation (E).
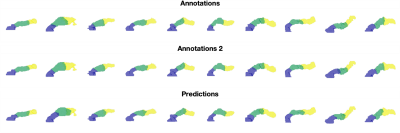
Figure 2. Variability in parts segmentation for the first 10 out of N=20 validation subjects for the first annotations (top), second annotations (middle), and our method’s predictions (bottom).
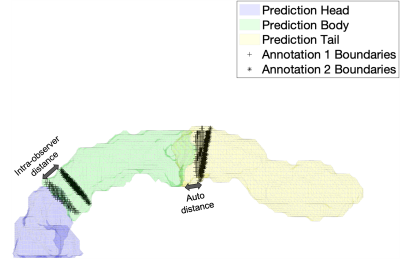
Figure 3. Illustration of the evaluation of parts segmentation using root mean squared error (RMSE) of the Euclidean distance between the annotation boundaries (black markers) and the predicted boundaries (solid colors). Illustrations for the intra-observer distance (left arrow), as well as distance between annotation 1 and prediction (right arrow), are shown.
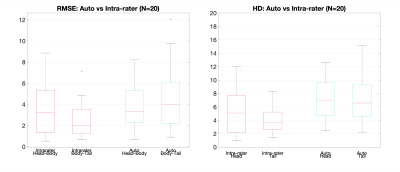
Figure 4. Validation of the method on 20 subjects using root mean squared error (RMSE) of the Euclidean distance between the 2 point clouds defined by the annotation boundary and the predicted boundary (left). We also reported Hausdorff distance (HD), recognising that only HD in head and tail are truly independent measurements (right). Intra-observer distance (annotation 1 vs annotation 2) was used as upper threshold performance.
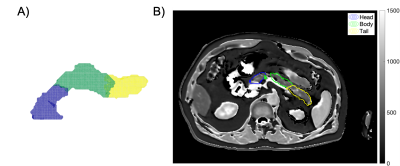
Figure 5. Automated parts segmentation of a given subject (A) enables automated regional quantification of pancreas imaging biomarkers, such as T1 (B). Example using a UK Biobank T1 map from a Shortened Modified Look-Locker Inversion recovery (ShMoLLI) scan. The resultant median values were 588 ms in the head, 556 ms in the body, and 566 ms in the tail.