1865
Evaluation of Modified Convolutional Neural Network for Automatic Measurement of Pancreas Volume and Pancreatic Fat Deposition1School of Biomedical Science, Auckland University of Technology, Auckland, New Zealand, 2Auckland University of Technology, Auckland, New Zealand, 3University of Auckland, Auckland, New Zealand, 4. Canterbury Health Laboratories, Christchurch, New Zealand, 5Saint Kentigern College, Auckland, New Zealand
Synopsis
Pancreatic fat has been reported to be closely related to type 2 diabetes risk, hence is the subject of our investigation in a clinical trial. Artificial pancreatic fat quantification is an experienced operator based and time consuming task. In our recent task, a convolutional neural network were trained based on latest accurate artificial quantification method. Result showed the identification rate were significantly improved through the program.
BACKGROUND
Accumulation of ectopic fat in the pancreas has been linked to type 2 diabetes risk1-3. Thus, quantification of such changes provides huge potential in terms of prognosis, diagnosis, and treatment to reduce metabolism disorders. However, the measurements of the pancreas size and fat percentage are usually time-consuming, and not available beyond experienced operators at academic centers. With the rapid development of Convolutional Neural Networks (CNNs), especially the U-Net, advanced medical image analysis4. U-Net achieved remarkable results on considerable medical image segmentation tasks to date in the heart, liver, kidney, and spleen. However, pancreas segmentation is still challenging due to high variability in size and positioning among patients 5. The relative softness of the pancreas made it easy to be squeezed by its surrounding organs. This also ambiguates the boundaries of the pancreas, which collapses with other non-pancreatic soft tissues, such as the small intestine, blood vessels, visceral adipose tissues. On the other hand, more information is shown from 3D MRI scans than 2D images, which makes it harder to establish complicated 3D models for context learning due to the limitation of current GPU memory size. In this paper, we established a coarse-to-fine 2D framework for auto pancreas segmentation based on the latest pancreatic fat calculation method, this resulted in improved accuracy of segmentation.METHODS
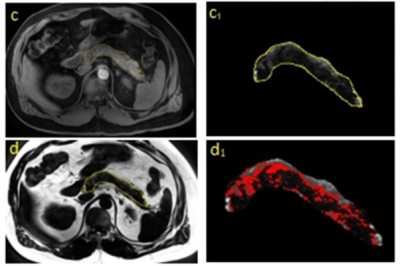
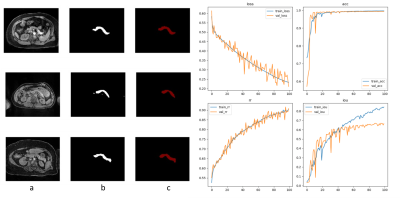
we established a modified coarse-to-fine 2D framework for auto pancreas segmentation, which includes two consecutive stages, localization stage, and refinement stage. The framework was designed on the basis of confronting artificial pancreatic fat segmentation. The data was from either patients or volunteers enrolled in our clinical research programs at Auckland Central Hospital. We totally used 120 patients for this study. Among those, 100 patients with approximately 1500 images were used for machine training, and the remaining 20 were used for validation. In order to solve the problem of the discontinuity of the grey pixel value and the ambiguity edge between pancreatic and other tissues in the localization stage, we enrolled superpixel segmentation, which is a shallow learning model that initially clusters image pixels based on their local structural features and spatial characteristics between them. This method aggregates image pixels into a series of adjacent pixel blocks with similar color, brightness, texture, which enhances the boundary contrast between superpixels. We also apply the erosion method to get rid of the impact of the boundary noise. The erosion of the boundary values was adjusted until CNN was trained to get a stable and satisfying value. The trained framework was then applied to measure a set of patients and the values were then compared with the artificial segmentation result.RESULTS
In our study, we established an improved U-NET network for pancreatic segmentation from MRI images. The U-NET was based on the accurate artificial segmentation method shown in Fig 1. combining the characteristics of superpixels and erosion boundaries which are shown in Fig 2. This method improved both the accuracies of pancreatic segmentation and the overall training speed of the network. The segmentation results were shown in Fig 3. We used the DSC value as the criterion of our training set and used a 4-fold cross-validation evaluation method on the NIH pancreatic segmentation dataset in this paper. Our experiments had shown that the average DSC value obtained by the method was 92.3%, which is 4.8% higher than that of normal U-NET. The results have also verified the effectiveness of our framework whose computing speed is faster than U-NET.DISCUSSION
This is the first study to train an auto convolutional neural network for pancreatic segmentation combining the ideas of the clinical segmentation method. We also involved the characteristics of superpixel and boundary value erosion system to deal with the trade-off between sensitivity and specificity both in the localization stage and refinement stage. A novel slice interaction network with a slice correlation module is built for multi-level slice communication. Furthermore, our framework achieved state-of-the-art performance in an open pancreas segmentation dataset and can be easily adopted to other organ segmentation tasks. In the future, more advanced techniques will be applied to our training set and hopefully will achieve more functions and accuracy for the framework. For example, deep Q-learning has shown its strength in detection tasks. Besides, generative models will be investigated for data augmentation to deal with the special textual caused by vessels, pancreatic duct, or steatosis. Also, we plan to apply more data from our program to make it easier to derive some academic conclusions in terms of pancreatic fat and metabolic diseases.CONCLUSION
MRI method is the only reliable tool to measure the pancreatic fat change in the clinic, which may provide type 2 diabetes risk indication. The measurement is a time consuming and experienced operator based clinical work. It now can become a faster way using AI to recognize the pancreatic fat changes and correlate them to metabolic disorders. This also provides a possibility that prognosis any latency diseases via software or online in the near futureAcknowledgements
No acknowledgement found.References
1. Heni M, Machann J, Staiger H, et al. Pancreatic fat is negatively associated with insulin secretion in individuals with impaired fasting glucose and/or impaired glucose tolerance: a nuclear magnetic resonance study. Diabetes/metabolism research and reviews. 2010;26(3):200-205.
2. Toledo-Corral CM, Alderete TL, Hu HH, et al. Ectopic fat deposition in prediabetic overweight and obese minority adolescents. The Journal of Clinical Endocrinology & Metabolism. 2013;98(3):1115-1121.
3. Singh RG, Yoon HD, Wu LM, et al. Ectopic fat accumulation in the pancreas and its clinical relevance: a systematic review, meta-analysis, and meta-regression. Metabolism. 2017;69:1-13.
4. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention: Springer 2015:234-241.
5. Zhou Y, Xie L, Fishman EK, et al. Deep supervision for pancreatic cyst segmentation in abdominal CT scans. International conference on medical image computing and computer-assisted intervention: Springer 2017:222-230.
Figures