1773
Noise2Recon: A Semi-Supervised Framework for Joint MRI Reconstruction and Denoising using Limited Data1Electrical Engineering (Equal Contribution), Stanford University, Stanford, CA, United States, 2Radiology, Stanford University, Stanford, CA, United States, 3Electrical Engineering, Stanford University, Stanford, CA, United States, 4Bioengineering, Stanford University, Stanford, CA, United States, 5Equal Contribution, Stanford University, Stanford, CA, United States, 6Biomedical Data Science, Stanford University, Stanford, CA, United States
Synopsis
Deep learning (DL) has shown promise for faster, high quality accelerated MRI reconstruction. However, standard supervised DL methods depend on extensive amounts of fully-sampled, ground-truth data and are sensitive to out-of-distribution (OOD), particularly low-SNR, data. In this work, we propose a semi-supervised, consistency-based framework (termed Noise2Recon) for joint MR reconstruction and denoising that uses a limited number of fully-sampled references. Results demonstrate that even with minimal ground-truth data, Noise2Recon can use unsupervised, undersampled data to 1) achieve high performance on in-distribution (noise-free) scans and 2) improve generalizability to noisy, OOD scans compared to both standard and augmentation-based supervised methods.
Introduction
Accelerated MRI can alleviate the high cost and slow speed of MR imaging. Current reconstruction methods rely on parallel imaging (PI) and compressed sensing (CS), but these methods have limited acceleration or require long reconstruction times1–3. Recent deep learning (DL) approaches may enable further accelerations compared to PI and CS4–6. However, standard supervised DL algorithms require vast amounts of fully-sampled references, which are difficult to collect prospectively at scale7. Additionally, the sensitivity of these algorithms to out-of-distribution (OOD) data, particularly low-SNR acquisitions, limit their generalizability8. Though supervised training on noise-augmented images is feasible, its impact on reconstructing both in-distribution (high-SNR) and OOD (low-SNR) data has not been investigated.While fully-sampled datasets are scarce, undersampled, noisy scans may be more accessible, leading to an opportunity to use both fully-sampled and undersampled scans in a semi-supervised DL approach. In this study, we propose Noise2Recon– a semi-supervised, consistency-based framework for joint MR reconstruction and denoising that uses a limited number of fully-sampled references. We evaluate whether Noise2Recon improves reconstruction performance and generalizability to noisy data in data-limited settings.
Theory
Noise2Recon uses denoising regularization to reduce overfitting to the image reconstruction task when supervised examples are limited. Unlike supervised algorithms, where only scans with fully-sampled references (supervised) are used during training, Noise2Recon training is complemented by a noise augmentation-consistency training paradigm (schematic-Fig.1). Examples without fully-sampled references (unsupervised) are augmented with noise, and the Noise2Recon model ($$$f_{\theta}$$$) generates reconstructions for both unsupervised images $$$x^{(u)}$$$ and noise-augmented undersampled images $$$x^{(u)} + \epsilon$$$. To encourage denoising during the reconstruction process, a consistency L1-loss is enforced between the two reconstructions (Eq.1), where $$$A$$$ is the imaging model and $$$i$$$ indexes the different coils. Here, noise is modeled as the complex-valued, masked noise: $$$\epsilon=UF\mathcal{N}(0,\sigma)$$$, where $$$U$$$ is the undersampling mask, $$$F$$$ is the Fourier operator, and $$$\mathcal{N}$$$ is a zero-mean complex-gaussian distribution with standard deviation σ. The total loss is the weighted sum of the supervised loss ($$$L_{sup}$$$) and the consistency loss ($$$L_{cons}$$$).$$\min_{\theta}\sum_i|f_{\theta}(x_i^{(u)},A_i)-f_{\theta}(x_i^{(u)}+\epsilon,A_i)|\;\;\;\;(1)$$
Methods
19 fully-sampled 3D fast-spin-echo knee scans (http://mridata.org) were partitioned into 14, 2, and 3 subjects (4480, 640, and 960 slices) for training, validation, and testing. 2D image reconstruction was performed for slices reformatted in the ky-kz direction. Data was undersampled using Poisson Disc undersampling with a 20x20 calibration region to estimate sensitivity maps using the JSENSE technique9. For each testing scan, a unique undersampling trajectory was generated using a reproducible random seed.To simulate reference-scarce settings, fully-sampled references were excluded for a random subset of training scans. For the training set $$$\mathcal{T}$$$, $$$\mathcal{T}_k\subset\mathcal{T}$$$ is the set of $$$k$$$ training scans for which fully-sampled references were retained. Scans not in $$$\mathcal{T}_k$$$ ($$$x\in\mathcal{T}\setminus\mathcal{T}_k$$$) were retrospectively undersampled with fixed masks to simulate unsupervised training scans. Experiments were performed at 12x and 16x acceleration factors for $$$k$$$=1,2,3,5,10 scans, where $$$\mathcal{T}_1\subset\mathcal{T}_2\dots\subset\mathcal{T}_{10}$$$.
To evaluate performance on noisy (OOD) test scans, fixed noise maps were generated at varying scan-magnitude-normalized standard deviations ($$$\sigma$$$=0.2,0.4,0.6,0.8,1.0) for each testing volume. During inference, these maps were added to the undersampled data following the noise model described above.
We compared Noise2Recon to two baselines: supervised training (Supervised) and supervised training with noise augmentations (Supervised+Aug). All DL networks used an unrolled architecture with eight residual blocks using the L1-loss10,11. Supervised baselines were only trained on examples with fully-sampled references ($$$x\in\mathcal{T}_k$$$). Noise standard deviation for Noise2Recon and Supervised+Aug was selected uniformly at random from a predefined range: $$$\sigma\in[0.2,1.0]$$$. Supervised+Aug augmentations were applied with a probability p=0.2 based on hyperparameter tuning. Image quality was measured using normalized root-mean-squared error (nRMSE), structural similarity (SSIM), and peak-signal-to-noise ratio (pSNR).
Results
With only one supervised scan ($$$k$$$=1), Noise2Recon had similar nRMSE and pSNR (Fig.2) on in-distribution scans compared to fully-trained ($$$k$$$=14) Supervised models, with example images shown in Fig.3. Supervised+Aug models consistently underperformed both Noise2Recon and Supervised baseline.Noise2Recon and Supervised+Aug models outperformed Supervised models on noisy scans (Fig.4). Unlike Supervised+Aug, Noise2Recon maintained high SSIM at varying noise levels with limited training data and recovered >95% of performance of the fully-trained Supervised+Aug model for all metrics (Fig.4). In noisy settings, Noise2Recon trained on a single acceleration generalized better to different accelerations (Fig.5).
Discussion
Noise2Recon achieved superior performance compared to both supervised baselines with limited data, particularly on noisy data. It also matched fully-supervised performance with almost 3x fewer examples. These results suggest that Noise2Recon may enable DL reconstruction with limited fully-sampled datasets and also provide a framework for using undersampled datasets during DL reconstruction.Expectedly, incorporating noise during training increased robustness for reconstructing noisy scans. However, standard noise augmentations on supervised scans (Supervised+Aug) were minimally effective with limited supervised data and degraded performance on in-distribution, high-SNR data. In contrast, Noise2Recon generalized well to varying extents of noise and accelerations even with limited data, while maintaining high performance on in-distribution data. This generalizability may enable low-SNR acquisitions (reduced NEX, increased resolution and readout bandwidth, lower field strengths, etc.) without compromising reconstruction fidelity. Simulating OOD data for consistency regularization beyond noise may also be beneficial to generalizing to other artifacts.
Conclusion
We presented Noise2Recon– a semi-supervised, consistency-based reconstruction and denoising method capable of achieving high performance on low-SNR, OOD examples without compromising performance on in-distribution data with limited supervised data.Acknowledgements
Research support provided by NSF 1656518, NIH AR077604, Precision Health and Integrated Diagnostics Center at Stanford, GE Healthcare, and PhilipsReferences
1. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: Sensitivity encoding for fast MRI. Magn Reson Med. 1999;42(5):952-962. doi:10.1002/(SICI)1522-2594(199911)42:5<952::AID-MRM16>3.0.CO;2-S
2. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182-1195. doi:10.1002/mrm.21391
3. Griswold MA, Jakob PM, Heidemann RM, et al. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA). Magn Reson Med. 2002;47(6):1202-1210. doi:10.1002/mrm.10171
4. Zbontar J, Knoll F, Sriram A, et al. fastMRI: An Open Dataset and Benchmarks for Accelerated MRI. 2018:1-29. http://arxiv.org/abs/1811.08839.
5. Chaudhari AS, Sandino CM, Cole EK, et al. Prospective Deployment of Deep Learning in MRI : A Framework for Important Considerations, Challenges, and Recommendations for Best Practices. J Magn Reson Imaging. August 2020:jmri.27331. doi:10.1002/jmri.27331
6. Sandino CM, Lai P, Vasanawala SS, Cheng JY. Accelerating cardiac cine MRI using a deep learning‐based ESPIRiT reconstruction. Magn Reson Med. 2021;85(1):152-167. doi:10.1002/mrm.28420
7. Cole EK, Pauly JM, Vasanawala SS, Ong F. Unsupervised MRI Reconstruction with Generative Adversarial Networks. arXiv. August 2020. http://arxiv.org/abs/2008.13065. Accessed December 12, 2020.
8. Knoll F, Hammernik K, Kobler E, Pock T, Recht MP, Sodickson DK. Assessment of the generalization of learned image reconstruction and the potential for transfer learning. Magn Reson Med. 2019;81(1):116-128. doi:10.1002/mrm.27355
9. Ying L, Sheng J. Joint image reconstruction and sensitivity estimation in SENSE (JSENSE). Magn Reson Med. 2007;57(6):1196-1202. doi:10.1002/mrm.21245
10. Diamond S, Sitzmann V, Heide Gordon Wetzstein F. Unrolled Optimization with Deep Priors.; 2018.
11. Sandino CM, Cheng JY, Chen F, Mardani M, Pauly JM, Vasanawala SS. Compressed Sensing: From Research to Clinical Practice with Deep Neural Networks: Shortening Scan Times for Magnetic Resonance Imaging. IEEE Signal Process Mag. 2020;37(1):117-127. doi:10.1109/MSP.2019.2950433
Figures
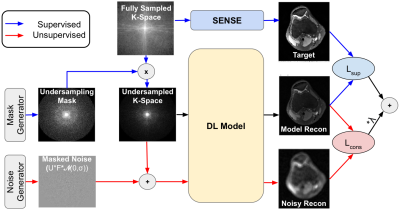
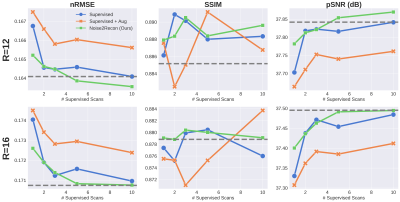
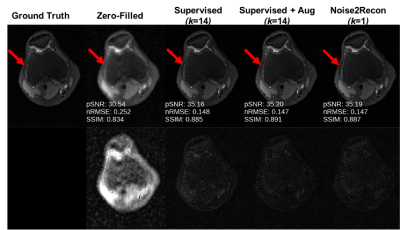
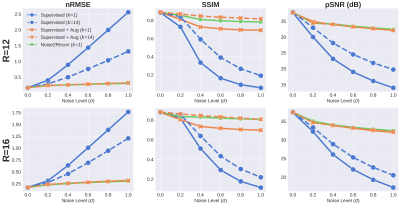
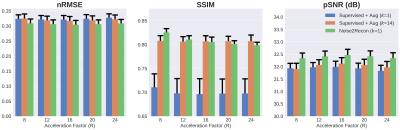
Fig. 5: Generalizing to reconstruction at multiple acceleration factors (R) at noise level σ=1 when trained on 16x undersampled images. k indicates number of supervised training scans. Noise2Recon outperforms Supervised+Aug with 7% of supervised data and reduces variance seen with supervised methods in the low-data regime. Additionally, Supervised+Aug baselines do not exhibit any performance recovery at lower acceleration factors (R=8,12). Noise2Recon improved performance in these cases without considerably compromising performance at higher accelerations (R=20,24).