1749
Adapting the U-net for Multi-coil MRI Reconstruction
Makarand Parigi1, Abhinav Saksena1, Nicole Seiberlich2, and Yun Jiang2
1Computer Science and Engineering, University of Michigan, Ann Arbor, MI, United States, 2Department of Radiology, University of Michigan, Ann Arbor, MI, United States
1Computer Science and Engineering, University of Michigan, Ann Arbor, MI, United States, 2Department of Radiology, University of Michigan, Ann Arbor, MI, United States
Synopsis
We propose a U-net based architecture for multi-coil MRI reconstruction. The model is able to utilize the multi-coil nature of the data by processing images before they are combined, unlike U-nets which require coil combination before inputting the image. We achieved SSIM scores higher than the U-net with much fewer parameters, enabling lower memory usage.
Introduction
Deep learning has been shown to be effective in reconstructing high quality MR images from undersampled k-space data.1, 2 One of the most popular models, the U-net, is able to reconstruct high quality images via an architecture that takes in a single image as input and outputs a refined image.3 We propose the Multinet, a U-net based architecture that operates in two places in the multi-coil reconstruction process: on the multi-coil images and on the single coil-combined image. In this way, the multi-coil nature of the data may be leveraged for reconstruction, enabling a parallel-imaging-like improvement in image quality. The Multinet performs better than a standard U-net when comparing evaluation metrics, while using 50% fewer parameters. A low parameter model means lightweight memory usage, allowing the model to be run effectively on more hardware.4Methods
The U-net employs a series of convolutions in an encoder-decoder architecture with skip connections.3 To apply a U-net to multi-coil data, the k-spaces from each coil are converted into images by applying the Inverse Fast Fourier Transform (IFFT). These images are then combined into a single image using Root-Sum-of-Squares (RSS), and the coil-combined image is refined by the U-net. The U-net does not take into account the coil sensitivity differences, which can supply additional information for reconstruction.The Multinet is composed of two U-nets: a U-net to operate on multiple images and a U-net to operate on a single image, as illustrated in Figure 1. The first U-net, called the Multi-image U-net, is a U-net with n input channels, where n is the number of receiver channels. It produces n images as output. The Multi-image U-net does not operate independently on each input image but as a whole, allowing the neural network to learn interdependencies between the input images. The Multi-image U-net is illustrated in Figure 2. The traditional U-net is a special case of this architecture with n=1.
A description of the reconstruction process with the Multinet follows, and is shown in Figure 1. The n undersampled k-spaces acquired from the MRI scanner are subjected to the IFFT to generate n aliased images. These n images are then input into the Multi-image U-net. An example of this multi-image refinement is shown in Figure 3. The resulting images are then combined into a single image using RSS. The single coil-combined image is then processed by a traditional U-net for further noise removal. The training loss is computed with an L1 function on the model reconstruction and the ground truth image, where the ground truth is generated with the fully sampled k-spaces and RSS.
In Souza et al., the authors evaluate the "W-net II," which is a cascade of two multi-image U-nets, and coil combination is done as the very last step.5 The Multinet differs from this architecture as it uses a single Multi-image U-net before combination, then further refines the combined image with a traditional U-net.
To produce U-nets of varying parameter sizes, the number of output channels of the first convolution layer is modified. Note that this is not necessarily the number of output channels of the entire model. To control the number of parameters of the Multinet model, the number of output channels of the first convolutions of each of the two component U-nets is modified.
The Multinet was trained on a subset of the fastMRI multi-coil brain dataset.5 As the model requires a constant number of coils, only those slices with 16 coils were included for training. Each model was trained on NVIDIA Tesla V100 GPUs at the University of Michigan's Great Lakes HPC cluster. The k-spaces were undersampled at a 4x acceleration rate with a random mask type with 0.08 center fractions. The training procedure used RMSProp gradient descent with a learning rate of 0.001. The quantitative results describe the model performance on the 16-coil subset of the fastMRI multi-coil brain validation dataset.
Results
Performance metrics of the Multinet are shown in Figure 4 and qualitative results on three particular slices of various acquisition types in Figure 5. The performance metrics include Structural Similarity Index Measure (SSIM), Normalized Mean Squared Error (NMSE), and Peak Signal-to-Noise Ratio (PSNR). "Unet-X" refers to a U-net where the first convolution layer has X output channels. Similarly, "Multinet-16" is a Multinet comprising a Multi-image Unet-16 and a Unet-16. The Multinet-16 uses 14.8MB of memory for its parameters while the Unet-32 uses 29.6MB, about double. The Multinet-16 achieves 0.9346 SSIM, greater than the Unet-32's 0.9276 SSIM. Similarly, it has a higher PSNR and a lower NMSE than the Unet-32. On the images in Figure 5, The Multinet-16 achieves a higher SSIM than the Unet-32 on both the T1 and T2 weighted examples, and is marginally lower on the FLAIR example.Discussion and Conclusion
These results suggest that information that may be helpful for reconstruction is lost when images are coil combined; unlike the IFFT, RSS is not invertible. Training models before and after the coil combination step of reconstruction may mean higher quality images with lower memory usage. Future work may involve testing various models before the coil combination step to optimize the pre-combination images.Acknowledgements
No acknowledgement found.References
- Schlemper, J., Caballero, J., Hajnal, J. V., Price, A., & Rueckert, D. (2017, June). A deep cascade of convolutional neural networks for MR image reconstruction. In International Conference on Information Processing in Medical Imaging (pp. 647-658). Springer, Cham.
- Wang, S., Su, Z., Ying, L., Peng, X., Zhu, S., Liang, F., ... & Liang, D. (2016, April). Accelerating magnetic resonance imaging via deep learning. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) (pp. 514-517). IEEE.
- Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
- Hyun, C. M., Kim, H. P., Lee, S. M., Lee, S., & Seo, J. K. (2018). Deep learning for undersampled MRI reconstruction. Physics in Medicine & Biology, 63(13), 135007.
- Souza, R., Bento, M., Nogovitsyn, N., Chung, K. J., Loos, W., Lebel, R. M., & Frayne, R. (2020). Dual-domain cascade of U-nets for multi-channel magnetic resonance image reconstruction. Magnetic resonance imaging, 71, 140-153.
- Zbontar, J., Knoll, F., Sriram, A., Murrell, T., Huang, Z., Muckley, M. J., ... & Parente, M. (2018). fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv preprint arXiv:1811.08839.
Figures

Figure 1: Illustration of the Multinet architecture, featuring the Multi-image U-net and a traditional U-net. The Multi-image U-net takes in as many images as there are coils, and outputs that many images.
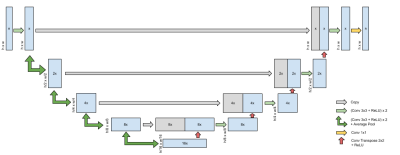
Figure 2: Illustration of the Multi-image U-net, the first component of the Multinet. The number in the boxes indicates the number of channels, while the width and height of the data is displayed to the side. x is the number of channels in the output of the first convolution, i.e. Multinet-16 would have x=16. n is the number of input and output channels of the entire model. For the Multinet, this is the number of coils.
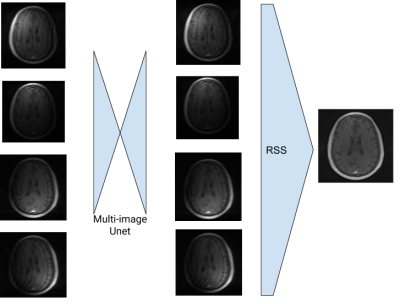
Figure 3: The Multi-image U-net performs noise reduction before the images are combined with RSS.
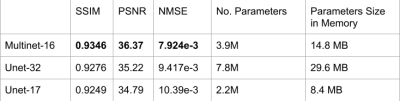
Figure 4: Performance on fastMRI multi-coil brain validation dataset, restricted to slices with 16 coils. In the Multinet-16, each component U-net has about 1.9M parameters, for a total of about 3.9M parameters. For SSIM and PSNR, higher is better; for NMSE, lower is better.
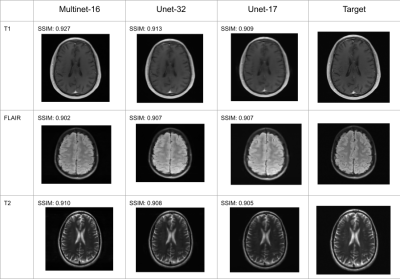
Figure 5: Reconstructions and SSIM scores of T1, FLAIR, and T2 weighted images.