1740
Proof-of-principal of 1H PREFUL approaches at 0.5T: lung ventilation and perfusion maps of a healthy volunteer and a Covid-19 survivor
Arthur Harrison1, Galina E Pavlovskaya1,2, Thomas Meersmann1, Olga S Pavlova3,4, Alexander Makurenkov4, Yury A Pirogov4, and Nikolay V Anisimov3,4
1SPMIC/Medicine, University of Nottingham, Nottingham, United Kingdom, 22Nottingham NIHR Biomedical Research Centre, Nottingham, United Kingdom, 3Faculty of Fundamental Medicine, Lomonosov Moscow State University, Moscow, Russian Federation, 4Faculty of Physics, Lomonosov Moscow State University, Moscow, Russian Federation
1SPMIC/Medicine, University of Nottingham, Nottingham, United Kingdom, 22Nottingham NIHR Biomedical Research Centre, Nottingham, United Kingdom, 3Faculty of Fundamental Medicine, Lomonosov Moscow State University, Moscow, Russian Federation, 4Faculty of Physics, Lomonosov Moscow State University, Moscow, Russian Federation
Synopsis
The ability to visualise non-invasively impaired lung ventilation and perfusion caused by lung scarring is important in monitoring recovery of Covid-19 survivors. The hp Xenon-129 imaging modality is currently approved for clinical use in the UK only, however, it has been shown recently that 1H PREFUL methodology can be used to visualise lung perfusion and ventilation in health and disease at 3T whole body scanners. However, 3T scanners may not be available for healthcare in countries with low and middle income, hence PREFUL at 0.5T was used to assess lung ventilation and perfusion in health and Covid-19 disease.
Introduction
The effects of Covid-19 on lungs have been recently demonstrated using hyperpolarised (hp) xenon imaging modality in the UK using 1.5T whole body MRI [1]. Hyperpolarised (hp) xenon imaging modality and 1.5T and 3T clinical scanners are not readily available for healthcare in countries with low and middle income hence the potential availability of low (< 1.5T) whole body 1H lung imaging modality that can assess ventilation and perfusion of lungs in health and disease is of great importance as low field strength scanners are abundantly available in healthcare systems in many middle and low income countries worldwide.Purpose
To use PREFUL methodology [2] to highlight the differences in ventilation and perfusion of lungs of a healthy volunteer and a Covid-19 survivor as a proof-of-principle at 0.5T.Methods
All experiments were carried out on a 0.5 T MR scanner Tomikon S50 (Bruker, Ettlingen, Germany located in Centre for Magnetic Resonance Imaging and Spectroscopy, Moscow State University, Moscow, Russia) equipped with a superconducting magnet (Magnex, Oxford, UK) with a bore diameter of 60 cm, a 2 kW RF transmitter LPPA 2120 (Dressler, Stolberg, Germany) and a S630 gradient system with a maximum power of 16.7 mT/m with a rise time of 0.5 ms. Scan management and primary data processing were performed using XWinNMR v.1 and ParaVision v.1. Subsequent data processing were performed using the open-source software package ImageJ. PREFUL algorithm was implemented in Matlab R2020a and used to reconstruct ventilation and perfusion maps. The build in 1H whole body coil was used in all experiments. Images were acquired using 2D GRE protocol with TR/TE=9.6/2.7 ms, BW=52.6 kHz, FOV=27.5×25 cm2, matrix size =88×80, slice thickness of 15 mm and acquisition time of 4 m 9 sec for 200 frames with 0.845s per frame, or TR/TE=3.21/2.14 ms, P(sinc3)=0.1 ms, BW=200 kHz, FOV=22×25 cm2, MTX=44×50, ST = 25 mm and acquisition time of 1 m 9s with for 200 frames with 0.341s per frame using free breathing in all experiments. One female (26y) healthy volunteer and one male Covid-19 survivor (40y) were recruited for this proof of principal study. All necessary ethical permits were obtained prior to the study.Results and Discussion
Selected images extracted from free breathing time series with 0.845 s per frame ST = 15mm for both healthy volunteer and a Covid-19 survivor with corresponding ventilation images are displayed in Figure 1. Selected images from the times series with 0.34s per frame are displayed in Figure 2. The shortening of the acquisition time per frame allowed for sampling of the cardiac cycle that was used to produce lung perfusion images. However, this resulted in the removal of slice selection in GE protocol, and therefore, in the loss of spatial resolution dictated by the demand to produce quality images suitable for further analysis. The resulting images are displayed in Figure 2. While ventilation images reconstructed using PREFUL algorithm are similar for both subjects, perfusion demonstrates defects in lungs of the Covid-19 survivor. This is probably associated with fibrosis in lungs induced by the aggressive course of Covid -19 in this subject. The subject will be scanned in one month time to characterize trajectory in perfusion and ventilation during their recovery period.Conclusion
PREFUL methods might be useful for Covid-19 lung recovery monitoring at 0.5T. This methodology should be extended to other pulmonary patient groups to check its efficacy. The implementation of this methodology in middle and low income countries might open low field MRI for the purpose of monitoring lung function in recovery and acute settings.Acknowledgements
The study was supported by RFBR grants 19-29-10015 and 20-52-10004. the Royal Society (Grant # EC\R2\192175).References
1. https://www.bbc.co.uk/news/health-55017301 , 2. Andreas Voskrebenzev , Marcel Gutberlet , Filip Klimeš , Till F Kaireit , Christian Schönfeld , Alexander Rotärmel , Frank Wacker, Jens Vogel-Claussen “Feasibility of quantitative regional ventilation and perfusion mapping with phase-resolved functional lung (PREFUL) MRI in healthy volunteers and COPD, CTEPH, and CF patients”, Magn Reson Med. 2018 Apr;79(4): 2306-2314, doi: 10.1002/mrm.26893 .Figures
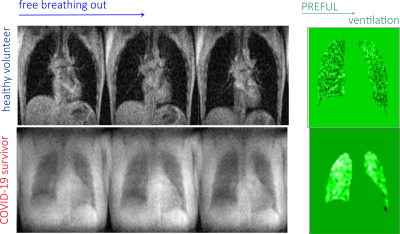
Figure
1. Selected time series and PREFUL lung ventilation images in
a healthy volunteer and Covid-19 survivor. Note that ventilation defects present
in the Covid lung are minor.
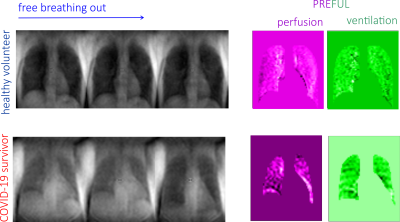
Figure
2. Selected time series and PREFUL lung ventilation
images in a healthy volunteer and Covid-19 survivor
with sampled cardiac cycle. Note that the ventilation and perfusion defects become
more apparent in the Covid lung.