1723
Semi-automated assessment of the principal diffusion direction in the corpus callosum: application across brain diseases1Neuroscience Research Center, University "Magna Graecia", Catanzaro, Italy, 2Institute of Neurology, University "Magna Graecia", Catanzaro, Italy
Synopsis
We propose a novel, semi-automated approach to assess corpus callosum integrity at the individual level, based on the spatial distribution of the principal diffusion direction orientation. We applied the method to the clinical challenge of differentiating normal pressure hydrocephalus (iNPH, N=23) from progressive supranuclear palsy (PSP, N=27) and Alzheimer’s Disease (AD, N=35), whose shared clinical and radiological features can make early differential diagnosis difficult. We found differential involvement of PDD distribution across neurodegenerative (AD, PSP) and non-degenerative diseases (such as iNPH), a finding that could shed light on the different mechanisms that underlie tissue damage.
Introduction
Different neurological disorders induce changes in the microstructural integrity of corpus callosum (CC) that can be investigated using diffusion-weighted MRI (dMRI). Most commonly used dMRI-derived scalars, such as mean diffusivity (MD) and fractional anisotropy (FA), however, lack specificity to underlying pathophysiology, and fail to consider the three-dimensional nature of diffusion, crucial for a reliable characterization of brain tissue microstructure. The orientation of the principal eigenvector of the tensor - i.e., principal diffusion direction (PDD) or V1 - has been investigated less, but could provide more information on the nature of tissue changes and help to identify promising markers that could be focus of further analysis using more accurate dMRI models. Here, we propose a novel, semi-automated approach to assess CC integrity at the individual level, based on the spatial distribution of PDD. We applied the method to the clinical challenge of differentiating normal pressure hydrocephalus (iNPH, N=23) from progressive supranuclear palsy (PSP, N=27) and Alzheimer’s Disease (AD, N=35), whose shared clinical and radiological features can make early differential diagnosis difficult.1 We also assessed the method’s generalizability over different scanners using multi-site dMRI data.Methods
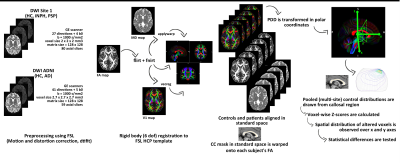
A detailed workflow of the proposed analysis is shown in Figure 1. Briefly, PDD maps were obtained from multi-site 3T dMRI data, appropriately aligned in standard space using a rigid-body registration – to avoid attempting a deformation procedure on very damaged brains. Eigenvectors at each voxel were then decomposed in polar coordinates (theta and phi angles). Subsequent analyses were restricted to the midsagittal region of the CC (± 6 slices from the midsagittal plane); this was done to increase both anatomical correspondence across different subjects and accuracy of PDD estimation in a region where the direction of fibers has been very well described.2 The reference PDD distribution in the CC was drawn from 36 healthy volunteers (14 from Site 1, 22 from ADNI data). All four groups were age- and sex-matched. In the CC of each patient, a voxel was considered altered if the z-scores of both angles exceeded 2.5 standard deviations above or below the controls’ mean. The distribution of altered voxels was obtained for each subject: i) along the latero-lateral axis (x voxel coordinate) of the three main CC subregions (splenium, body and genu); ii) along the anteroposterior (y voxel coordinate) profile of the midsagittal CC. To compare the behavior of PDD to that of the more common DTI-derived scalars, z-scores and distributions of voxels with "unusual" FA and MD values were also examined. Kruskal-Wallis tests followed by post-hoc pairwise comparisons were used to assess significant differences of the distribution values across groups. Statistical significance threshold was set at 0.05 after Bonferroni correction for multiple comparison.Results
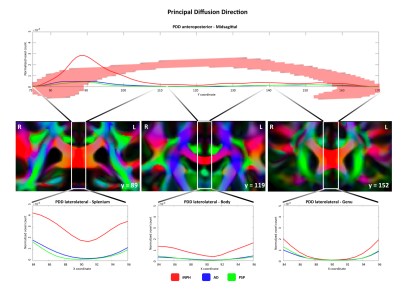
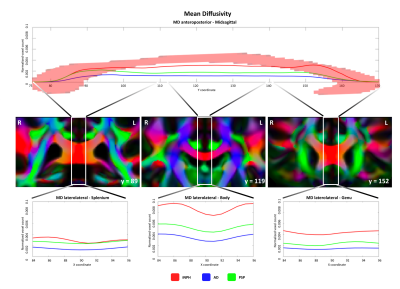
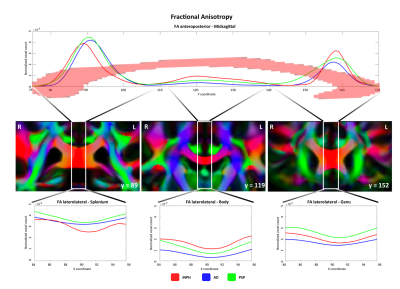
We observed different topographical distributions of changes in the CC: Figure 2 shows that patients with iNPH presented PDD alterations in the midsagittal portion of the splenium, which differed significantly from patients with AD and PSP. On the other hand, in Figure 3 is shown that the number of voxels with altered MD values was steady across the midsagittal profile, with iNPH and PSP significantly more affected than AD in the CC body and genu. Finally, Figure 4 shows that the FA profile was similarly altered across all three patients’ groups compared to the control distribution of FA values.Discussion
Alteration of PDD orientation in the CC splenium seems a promising correlate of disease in patients with iNPH but not in those with PSP and AD, a finding that was not detectable using only the classical scalar dMRI metrics. Interestingly, while FA and MD were altered in AD compared to controls, we observed that i) the orientation of the PDD was not altered in AD patients and ii) the extent of the region in which MD was altered in AD was significantly smaller compared to patients with iNPH and PSP. In the light of this, analysis of the differential involvement of PDD distribution across neurodegenerative (AD, PSP) and non-degenerative diseases (such as iNPH) could shed light on the different mechanisms that underlie tissue damage. It is possible, in fact, that while CC fibers are damaged in PSP and AD due to Wallerian degeneration, the frequently observed CC damage in iNPH could be due to the pressure that the ventricles exert on the bundle, “pushing” it upwards and subsequently deviating the principal direction of the callosal fibers from the physiological left-right direction.Conclusions
The proposed method provided novel information on iNPH-related tissue damage. The pipeline was successfully applied to multi-site data, and only involved rigid-body alignment to standard space, thus avoiding nonlinear deformations that would have been particularly challenging in patients with severely affected brains. Moreover, the approach is validated over single-shell data with a relatively low number of directions, to take into account its possible future application in routine clinical practice. In particular, future studies might focus on PDD distribution before and after shunting procedures (used to reduce cerebrospinal fluid pressure in iNPH) to test whether alterations are reversible and are associated with surgical outcome.Acknowledgements
No acknowledgement found.References
1. Di Ieva A, Valli M, Cusimano MD. Distinguishing Alzheimer's disease from normal pressure hydrocephalus: a search for MRI biomarkers. J Alzheimers Dis. 2014;38(2):331-50.
2. Aboitiz F, Scheibel AB, Fisher RS, et al. Fiber composition of the human corpus callosum. Brain Res 1992;598:143–153.
Figures