1696
A novel multi-filter convolutional neural network for prediction of cognitive deficits using structural connectome in very preterm infants1Department of Radiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Department of Electronic Engineering and Computing Science, University of Cincinnati, Cincinnati, OH, United States, 3Deep MRI Imaging Inc., Lewes, DE, United States, 4The Perinatal Institute, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States
Synopsis
We proposed a novel multi-filter convolutional neural network for prediction of cognitive deficits using brain structural connectome data. In contrast to 2D grid convolutional filters in traditional convolutional neural networks, our proposed model contains multiple vector-shape convolutional filters that can better extract the topological information from brain connectome. We demonstrated the ability of our model to learn hidden patterns from brain connectome data for prediction tasks. Our proposed model was able to identify infants at a high risk of cognitive deficits with an area under the curve of 0.78, exceeding the performance of other existing peer convolutional neural network methods.
INTRODUCTION
Up to 40% of very preterm infants (i.e., ≤31 weeks gestational age) are diagnosed with cognitive deficits at 2 years of age 1. Such neurodevelopmental deficits affect infants throughout life, thereby resulting in poor educational and social outcomes 2. Unfortunately, no robust prognostic screening methods are currently available following neonatal intensive care stay. Recently, deep learning models, especially convolutional neural networks (CNNs), have shown great promise in prediction tasks using medical imaging data 3-5. In this study, we developed a novel multi-filter CNN for prediction of cognitive deficits using brain structural connectome data in very preterm infants. Compare to traditional CNN models with 2D grid convolutional filters, our proposed CNN model utilizes multiple vector-shape convolutional filters that can better leverage topological locality in brain connectome data for learning tasks.METHODS
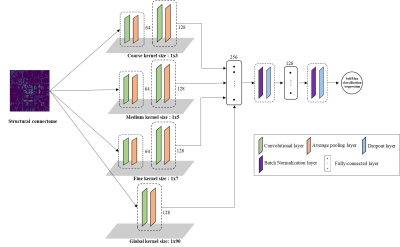
This study included 80 prospectively recruited very preterm infants with gestational age at birth mean (standard deviation) of 28 (2.4) weeks. The Nationwide Children's Hospital Institutional Review Board approved this study. Anatomical T2-weighted MRI and Diffusion tensor imaging (DTI) were performed at mean (SD) of 40 (0.6) weeks postmenstrual age. The details of the MRI scan parameters can be found in our previous work 3. All preterm infants received standardized Bayley Scales of Infant and Toddler Development III test at 2 years corrected age. The Bayley-III cognitive scores are on a scale of 40 to 160, with a mean of 100 and a standard deviation of 15. Using a cutoff value of <90, we dichotomized very preterm infants into groups of high-risk (<90) vs. low-risk for developing cognitive deficits. We preprocessed the obtained DTI data using FMRIB’s Diffusion Toolbox (in the FMRIB Software Library, FSL, Oxford, UK). Head motion and eddy current artifacts were mitigated by aligning all diffusion images to their B0 image via an affine transformation. We constructed the whole brain structural connectome based on 90 regions of interest (ROIs) defined from a neonatal Automated Anatomical Labeling (AAL) atlas 6. The obtained fractional anisotropy maps were harmonized using a batch-effect correction algorithm ComBat 7. The structural connectivity between each pair of ROIs was calculated as the mean fractional anisotropy of each voxel intersecting the tract and then averaged over all tracts between the two ROIs, resulting in a 90 x 90 symmetric adjacency matrix. This was performed using the UCLA Multimodal Connectivity Package 8.Our multi-filter CNN model (Figure 1) consisted of four convolutional channels with different sized vector-shape convolutional filters, ranging from coarse (1×3) and medium (1×5) to fine (1×7) and global (1×90). After each convolutional layer, an average pooling was applied. The outputs of four convolutional channels were flattened and concatenated, then connected to a single fully connected channel with two fully connected layers. We included batch normalization and dropout regularization layers after each fully connected layer. For risk stratification (i.e., two-class classification), we adopted the Sigmoid function in the output layer and binary cross-entropy as the loss function.
For cognitive score prediction (i.e., regression), we used linear function in the output layer and mean-squared error as the loss function. We validated the model using a 5-fold cross-validation (CV) with the metrics of accuracy, sensitivity, specificity, and area under the curve (AUC) for the risk stratification. We calculated Pearson’s correlation coefficient, mean absolute error (MAE) and standard deviation of the MAE for the regression of cognitive scores. We repeated such 5-fold CV 20 times and reported mean and standard deviation (SD).
RESULTS
Table 1 displays detailed demographics of enrolled subjects in this study. As shown in Table 2 and Table 3, our proposed multi-filter model was able to correctly identify at high-risk infants for cognitive deficits with an (mean ± SD) accuracy of 80% ± 4.8% and the Pearson’s correlation coefficient between the predicted and actual Bayley III cognitive scores was 0.38 ± 0.05 (p < 0.001). Compared with other peer convolutional methods, including traditional CNN 9, InceptionCNN 10 and BrainNetCNN 11, our proposed model achieved a higher accuracy by 8.2% (p<0.001), 7.6% (p<0.001), and 4.8% (p<0.001), respectively.DISCUSSION and CONCLUSION
We proposed a multi-filter CNN to predict cognitive deficits using brain structural connectome data. We constructed the brain connectome adjacency matrix based on a neonatal AAL brain atlas 6, resulting that spatially nearby regions were also adjacent on the adjacency matrix. Therefore, the grid locality embedded in the adjacency matrix may not directly correspond to the topological locality of the brain network. We designed vector-shape filter to better capture the brain connectome topological information from spatially related ROIs. In addition, instead of a single-size filter, we proposed to use multi-size filters, which can provide supplementary information for complementary understanding within different brain regions as well as across the entire brain. Last, compared with 2D grid filters, our proposed CNN with vector-shape filters is computationally efficient.Compared with existing peer CNN models, the proposed multi-filter CNN achieved improved performance in early prediction of cognitive deficit in very preterm infants. This current study can be considered as a proof-of-concept due to the limited sample size. A larger study with external validation is important to validate our approach to further assess its clinical utility.
Acknowledgements
This study was supported by the National Institutes of Health grants R21-HD094085, R01-NS094200, R01-NS096037, R01-EB029944, and a Trustee grant from Cincinnati Children’s Hospital Medical Center.References
1. Blencowe H, Cousens S, Oestergaard MZ, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. 2012;379(9832):2162-2172.
2. Rogers EE, Hintz SR. Early neurodevelopmental outcomes of extremely preterm infants. Semin Perinatol. 2016;40(8):497-509.
3. Chen M, Li HL, Wang JH, et al. Early Prediction of Cognitive Deficit in Very Preterm Infants Using Brain Structural Connectome With Transfer Learning Enhanced Deep Convolutional Neural Networks. Frontiers in neuroscience. 2020;14.
4. Chen M, Li H, Wang J, Dillman JR, Parikh NA, He L. A Multichannel Deep Neural Network Model Analyzing Multiscale Functional Brain Connectome Data for Attention Deficit Hyperactivity Disorder Detection. Radiology: Artificial Intelligence. 2019;2(1):e190012.
5. Li HL, Parikh NA, He LL. A Novel Transfer Learning Approach to Enhance Deep Neural Network Classification of Brain Functional Connectomes. Frontiers in neuroscience. 2018;12.
6. Shi F, Yap PT, Wu G, et al. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One. 2011;6(4):e18746.
7. Fortin JP, Parker D, Tunc B, et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage. 2017;161:149-170.
8. Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST. Conserved and variable architecture of human white matter connectivity. Neuroimage. 2011;54(2):1262-1279.
9. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436-444.
10. Szegedy C, Liu W, Jia YQ, et al. Going Deeper with Convolutions. 2015 Ieee Conference on Computer Vision and Pattern Recognition (Cvpr). 2015:1-9.
11. Kawahara J, Brown CJ, Miller SP, et al. BrainNetCNN: Convolutional neural networks for brain networks; towards predicting neurodevelopment. Neuroimage. 2017;146:1038-1049.
Figures