1609
Exploring parallel imaging performance for prostate imaging at 7T using a 72-channel receive array
Tijl van der Velden1, Mark Gosselink1, Ingmar Voogt2, Martijn Froeling1, Hans Hoogduin1, Dennis Klomp1, Bart Steensma1, and Alexander Raaijmakers1,3
1UMC Utrecht, Utrecht, Netherlands, 2Wavetronica, Utrecht, Netherlands, 3Biomedical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands
1UMC Utrecht, Utrecht, Netherlands, 2Wavetronica, Utrecht, Netherlands, 3Biomedical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands
Synopsis
In this study we investigate the parallel imaging performance of a 72 channel body array for 7 tesla. G-factor maps and T2w prostate images have been acquired in a healthy volunteer. Accelerations of 3x3 and 4x1 are feasible without detrimental g-factor penalties.
Introduction
Ultrahigh field MRI can profit from high SNR levels that can be interchanged with reduced scan time. However, this requires sufficient acceleration potential of the receive array setup. In fact, the potential for acceleration is larger anyway at 7T because the higher B1 frequency results in lower noise correlations and more distinct sensitivity patterns [1]. This calls for an exploration of acceleration performance with a massively parallel receive array. Several studies have been presented on massively parallel receive arrays and parallel imaging performance [2]. However, these studies have focused predominantly at the brain. In this study, we explore parallel imaging potential for body imaging at 7T using a 8 channel transceiver setup combined with a 64 channel receiver array (figure 1) [3]. The setup consists of 8 fractionated dipole antennas and 64 loops. In this study we have investigated the parallel imaging performance when using all 72 elements simultaneously for prostate imaging at 7T.Methods
The coil setup was connected to a 7 tesla whole body MR system (Philips, Best, NL; Software release 5.4), equipped with 8x2kW RF amplifiers, and 72 digital receivers for 1H. All of the experiments were performed in accordance with the local ethical committee, and written informed consent was obtained from a 33 year healthy male volunteer.To get an impression of the limits on the acceleration of parallel imaging, a multi-slice 2D PDw image was acquired to calculate 1/g-factor maps. Values below 0.5 in these maps indicate too much noise amplification. Based on the noise preparation data of the same scan, a noise correlation matrix was calculated to determine the coupling between all elements; the 8 antennas as well as the 64-channel receiver array.
To visualize the prostate and its structures, three 2D T2w TSE images were acquired, with SENSE accelerations of 1, 2 and 4 (RL direction). In addition, 2 3D T1w images were acquired, with SENSE accelerations of 1x1 and 3x3 (RLxFH direction) to demonstrate imaging performance on a larger area of the lower torso.
Results
Figure 2 shows the noise correlation matrix, where a correlation of typically 0.3 or lower is shown, indicating good decoupling between elements. Although several elements show a correlation between 0.4 and 0.5, little effect on the total performance is expected, due to the high number of receiver channels.This is reflected by the 1/g-factor maps, shown in figure 3 in transversal and coronal orientations. Accelerations up to a factor of 3 in any direction is expected to be feasible with little noise amplification.
Figure 4 shows the T2w images with SENSE acceleration 1, 2 and 4, and a close-up of the prostate. From an acceleration of 4 small unfolding artefacts can be observed, which was expected when analyzing the 1/g-factor maps. On top on that, the reduced SNR as a result of SENSE acceleration results in reduced contrast between tissue types.
Similar results can be seen in figure 5, which shows a video of the 3D T1w acquisition with SENSE accelerations of 1x1 and 3x3. Again, in accordance with g-factor maps, unfolding artefacts can be observed, predominately in peripheral regions of the body.
Conclusions
In this study we have explored the parallel imaging performance for prostate imaging with a 72-channel setup for body imaging at 7T. Accelerations up to 9 (3D acquisitions) and 4 (2D acquisitions) are shown to be feasible.Acknowledgements
No acknowledgement found.References
1. Wiesinger et al. MRM 2004, https://doi.org/10.1002/mrm.20183
2. Auerbach et al. ISMRM 2017, #1218
3. Voogt et al. ISMRM 2016, #0173
Figures

a)
16 elements around the body, alternating antenna combined with 4 loops, with an
element with 4 loops. b) two elements positioned around a phantom. c) complete
setup. d) Single receive loop (from Voogt et al).
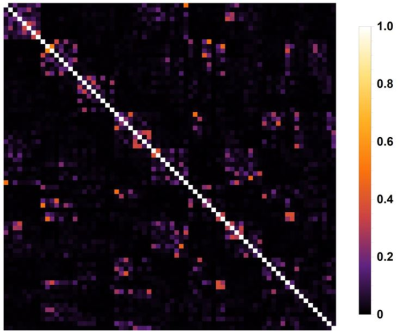
Noise correlation matrix with predominately values below 0.3,
indicating good decoupling.
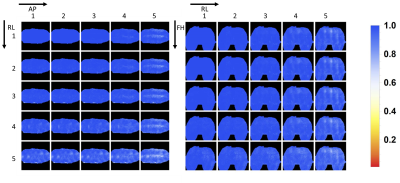
1/g-factor
maps. Values above 0.5 are considered good (equals g-factor < 2). Good acceleration
performance is particularly expected in the FH-direction and the LR-direction,
up to a factor of 3 in any direction.
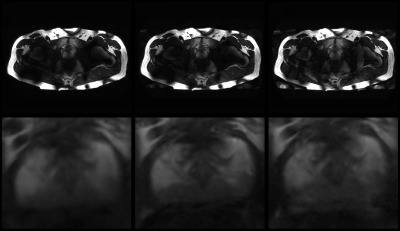
T2w
images with SENSE 1 (left), 2 (center) and 4 (right). Bottom row shows
magnification of the prostate. From an acceleration of 4 small artefacts are
observed, as well as the expected reduction in SNR.
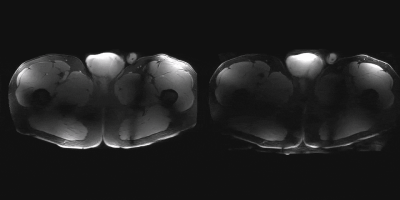
3D
T1w acquisition of the lower body. Left, no SENSE acceleration, right: 3x3
SENSE acceleration. Artefacts are predominately in the peripheral region of the
body.