1607
Development of a multiplexed ERETIC-RF array coil for quantitative whole brain 3D-MR spectroscopic imaging (MRSI)1Dept. of Radiology, MGH, A. A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Mittelhessen University of Applied Science, Giessen, Germany, 4Siemens Medical Solutions USA, Charlestown, MA, United States, 5Siemens Healthcare, Erlangen, Germany
Synopsis
An Electronic REference To access In vivo Concentrations (ERETIC) to absolutely quantify the brain metabolites for 3D-MRSI using an array coil was developed. The challenge of ERETIC for large receive arrays is that the addition of many ERETIC channels might negatively impact B0 and B1 homogeneity and increase the unwanted channel cross-talk. Here we investigated whether the number of ERETIC channels can be reduced below the number of RF receive elements. We also developed a unique method to coil combine the ERETIC and receive array signal. Metabolite concentration maps obtained with ERETIC were compared to internal water reference method.
INTRODUCTION
Magnetic Resonance Spectroscopy (MRS) can be used for the quantitative measurement of various metabolites in the brain non-invasively. The metabolite quantification can be achieved either by using internal water reference (IWR) or external reference methods such as phantom replacement and ERETIC (Electronic REference To access In vivo Concentrations) methods1. Using the IWR method for diseased conditions may introduce an error in the quantification as water content varies due to inflammation, hemorrhages, sclerosis, demyelination, edema, and cell death2. While, in the phantom replacement method, the phantom has to be adjusted to match with each in vivo coil loading and measured each time the subject is measured3. To mitigate these problems the ERETIC method has been used in which a pre-calibrated low power reference signal is injected into the RF coil via induction. In this work, we integrated the ERETIC method with 3D MRSI which simplifies the need for encoding the ERETIC signal in MRSI voxels4 and does not need to pre-distort the ERETIC signal to compensate for eddy current correction5. Furthermore, we aimed to reduce the number of ERETIC channels per receive element in order to reduce unwanted effects on B0 and B1 created by ERETIC hardware, and to minimize channel cross-talk.METHOD
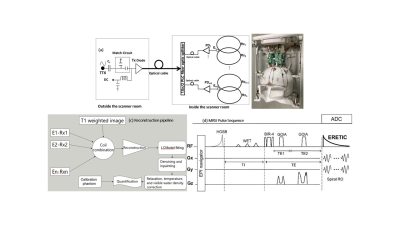
a) Hardware: An 8-channel RF receive array was integrated with ERETIC hardware. A schematic diagram is shown in Fig. 1a, in which synthetic signal generated by the RF synthesizer modulated to light using a laser diode [Tx] of wavelength 1300nm was carried via optical cable to a 4-way fiber optic splitter connected to 4 photo-diodes and further by double shielded RF coax cables to the micro-coils inductively coupled to receive loops. The geometry of the 8 receive loops allowed 1 ERETIC microcoil to be coupled inductively to 2 RF receive loops, hence the number of ERETIC channels was half of RF receive elements (Fig 1b). The optical path with minimum coax cables were used to minimize the parasitic coupling of ERETIC signal with other RF hardware and maximize inductive coupling with the RF coil which is key for ERETIC method6.b) MRSI sequence and reconstruction: An adiabatic spiral 3D MRSI sequence7 as shown in Fig. 1d with TR/TE = 1800/97 ms, FOV = 240X240X120 mm3, and matrix 34X34X9 was used to acquire metabolite and water reference data. The ERETIC signal injected in each receive element was measured separately as 1s pres-scan by an FID sequence that played the ERETIC pulse. This approach does not require eddy current predistortion of the ERETIC waveform. Acquired data were processed by the pipeline in Fig 1c: 1) ERETIC signal of each coil was used to scale the weights used during coil combination of raw MRSI data; 2) coil combined MRSI data were reconstructed and fitted with LCModel; 3) quality of metabolic maps was improved and outlier voxels with insufficient quality (CRLB > 20%, LW>0.1 ppm and SNR<3) were discarded followed by inpainting and denoising8 to fill the missing gaps; 4) for absolute quantification metabolite maps were voxel-wise corrected for T1 and T2 relaxation, WM, GM, and CSF segmentation and visible water content. The absolute concentration of metabolites was then computed using equations given in5. Brain segmentation was performed in FSL based on MEMPRAGE images.
c) Human subjects: Five healthy volunteers (4 males, 1 female, 30-35 years) were scanned with IRB approval.
RESULTS
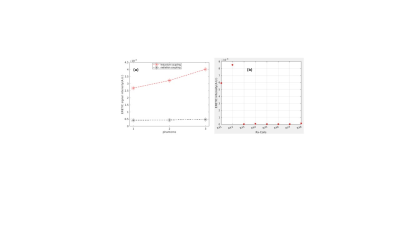
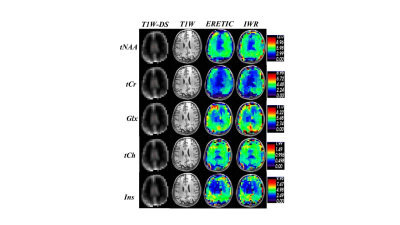
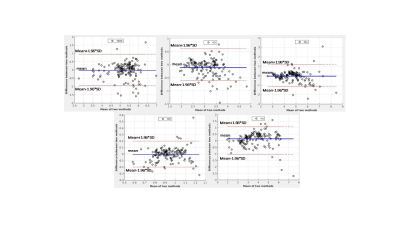
The performance of the ERETIC coil was evaluated by measuring the scaling of RF coil with different phantoms (Fig2a) and the response of RF loop to the ERETIC signal injected into the other RF loop(Fig.2b). In Fig. 2a the ERETIC signal (red) received with ERETIC coils placed near to the RF coil is scaled with different loading condition, while in radiation coupling (black) in which an ERETIC coil is placed at ~ 30 cm away from the RF coil, the signal does not change with loading. The received signal, in the latter case, is mainly due to the parasitic coupling of the micro-coil with RF coax cable and the pre-amplifier. The signal received by individual RF loops when an ERETIC signal was injected into the RF coils 1 and 2 is illustrated in Fig.2b which indicates that the influence of micro-coil on the non-adjacent RF coils is small. Fig.3 shows molar concertation maps of 5 representative brain metabolites: total NAA (tNAA), total creatine(tCr), glutamin and glutamate(Glx), total choline(tCh), and myo-inositol(Ins) obtained from ERETIC (3rd column) and IWR (4th column) method corresponding to the anatomical T1-weighted image in the 2nd column and down-sampled T1-weighted image in the 1st column. The Bland-Altman plots corresponding to these concertation maps are shown in Fig. 4 which indicates the good agreement between IWR and ERETIC methods for majority of the voxels.DISCUSSION
In this work we presented the ERETIC method of quantification for 3D-MRSI using array coil with ½ ratio between the number of ERETIC and RF channels. We addressed the challenges of ERETIC use in MRSI by eliminating the need for predistortion and simultaneous encoding of ERETIC and MRSI. Work is in progress to evaluate the performance of our approach in more healthy volunteers and patients.Acknowledgements
This work is supported by NIH (1R01CA211080-02) and MGH ECOR fund.References
1. Near J, Harris AD, Juchem C, et al. Preprocessing, analysis and quantification in single‐voxel magnetic resonance spectroscopy: experts’ consensus recommendations. NMR in Biomedicine 2020:1–23
2. Duc CO, Weber OM, Trabesinger AH, Meier D, Boesiger P. Quantitative 1H MRS of the human brain in vivo based on the simulation phantom calibration strategy. Magnetic Resonance in Medicine 1998;39:491–496. 3. Buchli R, Boesiger P. Comparison of methods for the determination of absolute metabolite concentrations in human muscles by 31P MRS. Magnetic Resonance in Medicine 1993;30:552–558.
4. Thapa B, A. Mareyam A, Stockmann J, Keil B, Hoecht P, Wang Z, Chang Y, Carp S, Li X, Strasser B, Wald L, Andronesi O, Towards absolute quantification of brain metabolites using Electronic REference To access In vivo Concentrations (ERETIC) for MR spectroscopic imaging (MRSI), ISMRM Conf. Proc., May 2019.
5. Zoelch N, Hock A, Heinzer-Schweizer S, Avdievitch N, Henning A. Accurate determination of brain metabolite concentrations using ERETIC as external reference. NMR in Biomedicine 2017;30:1–16.
6. Marro KI, Lee D, Shankland EG, et al. Synthetic signal injection using inductive coupling. Journal of Magnetic Resonance 2008;194:67–75.
7. Esmaeili M, Bathen TF, Rosen BR, Andronesi OC. Three-dimensional MR spectroscopic imaging using adiabatic spin echo and hypergeometric dual-band suppression for metabolic mapping over the entire brain. Magnetic Resonance in Medicine 2017;77:490–497.
8. Li X, Strasser B, Jafari-Khouzani K, Thapa B, Small J, Cahill D, Batchelor T, Andronesi O, Super-resolution 3D MRSI for Mapping 2HG and Tumor Metabolism in Patients with Mutant IDH Glioma, Neuroradiology, Jan2020.
Figures