1535
5-Minute MR Fingerprinting from Acquisition to Reconstruction for Whole-Brain Coverage with Isotropic Submillimeter Resolution1University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2Case Western Reserve University, Cleveland, OH, United States
Synopsis
Existing techniques for 3D MR fingerprinting focus on accelerating either acquisition or template matching, with typically limited spatial resolution. We develop a 3D MRF sequence coupled with a novel end-to-end deep learning image reconstruction framework for rapid and simultaneous whole-brain quantification of T1 and T2 relaxation times with isotropic submillimeter spatial resolution. We demonstrate feasibility with both retrospectively and prospectively accelerated 3D MRF data.
Introduction
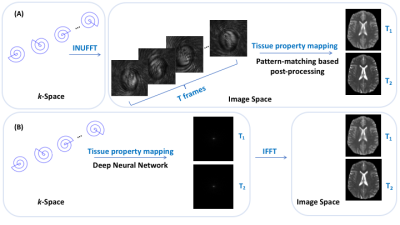
MR Fingerprinting (MRF) [1] allows measurement of multiple tissue properties in a single acquisition. Compared with 2D MRF, 3D MRF provides whole-brain coverage with higher spatial resolution and SNR. However, 3D MRF suffers from lengthy acquisition and processing times, limiting its clinical applicability. In this work, we develop (i) a 3D MRF sequence for isotropic submillimeter spatial resolution, and (ii) a novel end-to-end deep learning approach to infer multiple tissue properties directly from highly-undersampled spiral k-space MRF data, thereby avoiding time-consuming processing such as the non-uniform Fast Fourier Transform (NUFFT) and dictionary-based fingerprint matching. Experiments demonstrate that whole-brain 3D MRF acquisition and quantitative T1 and T2 mapping with 0.8 mm isotropic resolution can be achieved in a total time of ~ 5 mins.Methods
All MRI experiments were performed on a 3T Siemens scanner with a 32-channel head coil. Details on the 3D MRF pulse sequence are described in [2]. The 3D dataset was acquired using a stack-of-spirals trajectory. To achieve sub-millimeter isotropic resolution, a new spiral trajectory was designed with a maximum gradient magnitude of 32 mT/m and a maximum slew rate of 180 mT/m/msec. Only one spiral arm with a readout duration of 9.1 msec was acquired for each partition, corresponding to a high in-plane undersampling factor of 32. Other imaging parameters: FOV, 25×25 cm; matrix size, 320×320; slice thickness, 0.8 mm; number of slices, 120; GRAPPA factor along slice-encoding direction, 2; partial Fourier along slice-encoding direction, 6/8; flip angles, 5°~12°; MRF time points, 768; acquisition time, 18 min. To obtain the reference T1 and T2 maps for network training, non-Cartesian GRAPPA reconstruction was first applied along the slice-encoding direction, followed by NUFFT and dictionary matching (DM) using all 768 time points. Retrospectively undersampled 3D MRF data with R=4 along the slice-encoding direction and 25% of the total time points (192 time points) were used in training. Four subjects were used for training and one for testing. A 3D MRF dataset with prospectively undersampling (R=4 along slice-encoding direction and 192 time points) were collected for one subject. The acquisition time for whole-brain coverage (14 cm with sagittal slices) was ~4.8 min.The original MRF framework involves a two-step process of first reconstructing a series of time frames from non-Cartesian spiral k-space data using NUFFT, and then DM using the reconstructed time frames to infer tissue properties. In contrast, our method learns end-to-end a direct mapping from the acquired spiral k-space data to the tissue property maps (Fig 1). We demonstrate that gridding (core of NUFFT) and tissue property mapping can be performed simultaneously via a single deep neural network (e.g., U-Net [3]). Density compensation is achieved in the network via weighting as a function of the locations of data points on the k-space sampling trajectory. Tissue property mapping is carried out using a U-Net. Once trained, the network can directly consume the non-Cartesian k-space data, perform adaptive density compensation, and predict multiple tissue property maps in one forward pass (~0.34s per slice).
Results
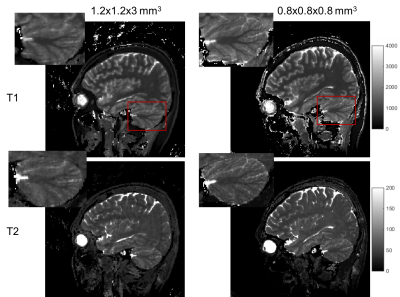
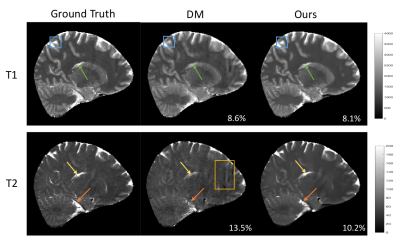
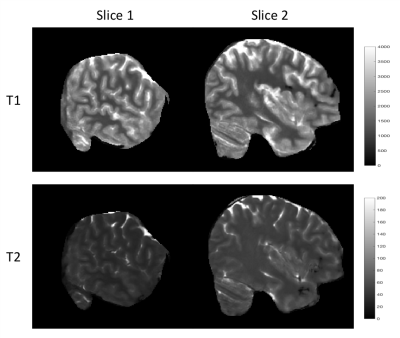
Fig. 2 shows an example of the high-resolution tissue property maps obtained using our 3D MRF sequence, demonstrating marked improvement in details over the 1.2x1.2x3 mm^3 resolution. Fig 3 shows that our method outperforms DM both qualitatively and quantitatively. Particularly, the maps obtained with our method do not contain the aliasing artifacts exhibited in the maps obtained with DM. The T1 and T2 voxel-average percentage mean absolute errors (MAE) are 11.1% and 15.7% for our method, reduced from 16.8% and 26.5% for DM. Our end-to-end tissue property mapping (~0.34s per slice) is ~1100 times faster than the original NUFFT-DM scheme. Fig. 4 shows representative maps obtained from a prospectively accelerated acquisition, demonstrating the feasibility of our method in rapid whole-brain quantitative imaging.Discussion
Our results indicate that acceleration along the slice-encoding direction in 3D MRF poses challenges for dictionary matching especially at high spatial resolution. In contrast, our method improves tissue quantification accuracy, likely due to the end-to-end optimization, directly linking the k-space data to the tissue maps. Rapid tissue mapping (~0.34s per slice) is important in clinical workflows, allowing timely re-scan decisions to be made without having to reschedule additional patient visits. Our method is also flexible as it allows any 2D deep neural network to be applied. Beyond MRF, the proposed framework can be easily adapted to facilitate non-Cartesian reconstruction in other applications.Conclusion
We proposed a novel, versatile and scalable approach for rapid high-resolution 3D MRF acquisition and tissue mapping. Experiments using accelerated MRF data with whole-brain coverage demonstrate that our method outperforms the conventional NUFFT-DM approach both in terms of accuracy and speed. In the future, we will train our network with a larger sample and will replace the U-Net network backbone with a more advanced network to further boost tissue quantification accuracy.Acknowledgements
This work was supported in part by NIH grant EB006733.References
[1] Ma, D., Gulani, V., Seiberlich, N., Liu, K., Sunshine, J. L., Duerk, J. L., & Griswold, M. A. (2013). Magnetic resonance fingerprinting. Nature, 495(7440), 187-192.
[2] Chen, Y., Fang, Z., Hung, S. C., Chang, W. T., Shen, D., & Lin, W. (2020). High-resolution 3D MR Fingerprinting using parallel imaging and deep learning. NeuroImage, 206, 116329.
[3] Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
Figures