1525
Oxytocin function regulation in autism spectrum disorders probed with rsfMRI complexity analysis1Emory University, Atlanta, GA, United States
Synopsis
Autism spectrum disorders (ASD) are characterized by dysregulation of endogenous oxytocin (OXT) function. In this study, we employed complexity of resting state fMRI signal as a tool to examine: 1) brain function impairments in adult ASD, 2) the relationship between DNA methylation of OXT receptor gene and severity of cognitive impairment, and 3) the effects of intranasal OXT treatment (IN-OXT). ASD patients exhibited abnormally increased fMRI signal complexity in a number of brain function networks. The severity of these impairments were inversely related to DNA methylation levels. IN-OXT attenuated impairments in these networks. Thus, OXT has neuroprotective effects in ASD.
INTRODUCTION
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by impairments in social interactions and communication, as well as repetitive behaviors and restricted interests 1. ASD is characterized by deficits in social cognition, face processing, and emotional regulation functions 1-3. Dysregulation of endogenous oxytocin (OXT) function has been reported in ASD 4, and its exogenous application was shown to enhance social functioning in ASD 5. ASD is characterized by disruptions in the brain’s excitation/inhibition (E/I) balance 6. OXT is also thought to enhance social cognition through its effects on GABA mediated control of E/I balance 6. Recently, an fMRI time-series signal complexity metric, multi-scale entropy (MSE) has been proposed as a potential biomarker of this E/I balance and neural activity 7-9. In this study, we employed MSE of resting state fMRI (rsfMRI) data to examine brain function impairments in ASD, and the effects of intranasal OXT (IN-OXT) treatment on ASD. Finally, OXT acts through the OXT receptor (OXTR) which is synthesized by the OXTR gene 10. The expression of OXTR gene across the brain is influenced by DNA methylation of that gene 10. DNA methylation levels of OXTR gene are an indication of endogenous OXT activity 10. Hence, we also investigated the relationship between DNA methylation of the OXT receptor gene and brain function impairments in ASD.METHODS
Thirty adult males with ASD (mean age ~ 29 years) were administered OXT and placebo intranasally in separate MR imaging sessions, in a double-blind study. For each ASD subject DNA methylation levels were obtained at six sites of the OXTR gene that were most implicated in ASD 11. Then ASD subjects and 17 age matched healthy controls (HC) who were not administered IN-OXT or placebo, were scanned in a Siemens 3T MRI Prisma-Fit scanner using a 64-channel Rx head+neck coil. Written informed consent was obtained from all participants in the protocol approved by the local Institutional Review Board. RsfMRI data were acquired with a 8-min whole-brain gradient echo EPI (TR/TE/FA = 3000ms/25ms/90°, resolution = 1.5mm x 1.5mm x 2.4mm). RsfMRI preprocessing steps included attenuation of signal related to subject-motion and physiological responses, using the AROMA techniques 12, spatial normalization to MNI152 space, and spatial smoothing with FWHM = 6mm isotropic Gaussian kernel. For each voxel time-series, sample entropy (SE) was calculated at 10 temporal scales using the Complexity toolbox 7,8, with window-length to m = 2, and distance threshold r = 0.3 (after optimization of parameters7,8). ROI-averaged MSEs were obtained from each brain resting state network (RSN) estimated from 1) Group ICA of ASD subjects’ placebo scan and HC; and 2) Group ICA of ASD subjects’ four fMRI scans at 48 IU, 24 IU, 8 IU doses of IN-OXT, and placebo. Between-group, and between-session differences in complexity were obtained with separate 2-sample (and paired) t-tests on each ICA RSN’s ROI averaged MSE. The relationship between the functional connectivity of each RSN with ASD subjects DNA methylation levels for each OXTR gene site were assessed with multiple linear regression.RESULTS & DISCUSSION
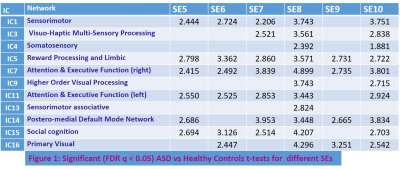
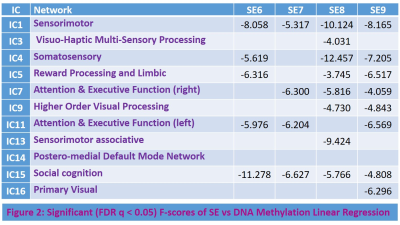
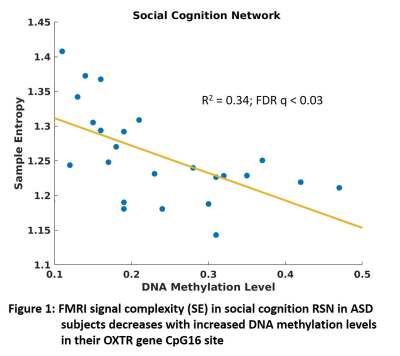
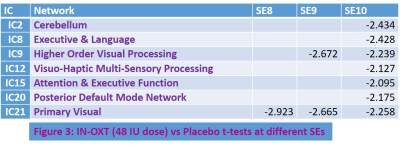
ASD patients exhibited (Table 1) significantly (FDR q < 0.05) abnormally increased SE compared to HC at longer temporal scales (5-10) in a number RSNs including social cognition, reward processing, limbic, attention, executive function, sensorimotor, and higher order visual processing networks. This is consistent with deficits in these functions seen in ASD 13-16. The abnormally increased complexity in these brain function networks could reflect increased synaptic noise arising from synaptic pruning deficits that are known to afflict ASD 17. Interestingly, the fMRI signal complexity in these RSNs were significantly (FDR q < 0.05) related to DNA methylation at the CpG16 site of OXTR gene (Table 2: Fig. 1). ASD subjects with increased DNA methylation levels exhibited less complexity (i.e., less synaptic noise) in brain function networks. This indicates that higher DNA methylation level at CpG16 has a neuroprotective effect on ASD. Finally, the 48 IU dose of IN-OXT significantly (FDR q < 0.05) reduced complexity at the long scales in a number of brain RSNs that exhibited abnormally increased complexity described above. This could indicates that the improvements in brain functional status in ASD in response to OXT 5 could arise from increased inhibitory control that attenuates synaptic noise in impaired brain function networks. Consistent with this notion, OXT has been shown to affect GABA-mediated E/I control 6.CONCLUSION
The results from this study implicate disturbances in E/I balance (due to increased synpatic noise potentially caused by synaptic pruning deficits) in a number brain function networks incuding those subserving social cognition and reward processing in ASD. OXT seems to attenuate the synaptic noise and hence likely normalize E/I balance. The results also indicate that fMRI signal complexity measures like MSE could act as diagnostic and therapeutic biomarkers of ASD.Acknowledgements
This work was supported by NIH grants P50MH100023, ORIP/OD P51OD011132, and UL1TR002378; as well as a pilot grant from Emory University Center for Mind, Brain, and Culture.References
1. Arnold Anteraper S, Guell X, D'Mello A, Joshi N, Whitfield-Gabrieli S, Joshi G. Disrupted Cerebrocerebellar Intrinsic Functional Connectivity in Young Adults with High-Functioning Autism Spectrum Disorder: A Data-Driven, Whole-Brain, High-Temporal Resolution Functional Magnetic Resonance Imaging Study. Brain Connect 2019;9:48-59.
2. Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods 2011;8:665-70.
3. Foss-Feig JH, Adkinson BD, Ji JL, et al. Searching for Cross-Diagnostic Convergence: Neural Mechanisms Governing Excitation and Inhibition Balance in Schizophrenia and Autism Spectrum Disorders. Biol Psychiatry 2017;81:848-61. 4. Yamasue H, Domes G. Oxytocin and Autism Spectrum Disorders. Curr Top Behav Neurosci 2018;35:449-65.
5. Ooi YP, Weng SJ, Kossowsky J, Gerger H, Sung M. Oxytocin and Autism Spectrum Disorders: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Pharmacopsychiatry 2017;50:5-13.
6. Lopatina OL, Komleva YK, Gorina YV, et al. Oxytocin and excitation/inhibition balance in social recognition. Neuropeptides 2018;72:1-11.
7. Wang DJJ, Jann K, Fan C, et al. Neurophysiological Basis of Multi-Scale Entropy of Brain Complexity and Its Relationship With Functional Connectivity. Front Neurosci 2018;12:352.
8. Smith RX, Yan L, Wang DJ. Multiple time scale complexity analysis of resting state FMRI. Brain Imaging Behav 2014;8:284-91.
9. McDonough IM, Nashiro K. Network complexity as a measure of information processing across resting-state networks: evidence from the Human Connectome Project. Front Hum Neurosci 2014;8:409.
10. Maud C, Ryan J, McIntosh JE, Olsson CA. The role of oxytocin receptor gene (OXTR) DNA methylation (DNAm) in human social and emotional functioning: a systematic narrative review. BMC Psychiatry 2018;18:154.
11. Andari E, Nishitani S, Kaundinya G, et al. Epigenetic modification of the oxytocin receptor gene: implications for autism symptom severity and brain functional connectivity. Neuropsychopharmacology 2020;45:1150-8.
12. Pruim RH, Mennes M, van Rooij D, Llera A, Buitelaar JK, Beckmann CF. ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage 2015;112:267-77.
13. Craig F, Margari F, Legrottaglie AR, Palumbi R, de Giambattista C, Margari L. A review of executive function deficits in autism spectrum disorder and attention-deficit/hyperactivity disorder. Neuropsychiatr Dis Treat 2016;12:1191-202.
14. Levy SE, Mandell DS, Schultz RT. Autism. Lancet 2009;374:1627-38.
15. Murphy E, Benitez-Burraco A. Language deficits in schizophrenia and autism as related oscillatory connectomopathies: An evolutionary account. Neurosci Biobehav Rev 2017;83:742-64.
16. Posar A, Visconti P. Sensory abnormalities in children with autism spectrum disorder. J Pediatr (Rio J) 2018;94:342-50.
17. Tang G, Gudsnuk K, Kuo SH, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014;83:1131-43.
Figures