1362
Learning-based automatic field-of-view positioning for fetal-brain MRI1Department of Radiology, Harvard Medical School, Boston, MA, United States, 2Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 3Computer Science and Artificial Intelligence Laboratory, MIT, Cambridge, MA, United States, 4Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children's Hospital, Boston, MA, United States
Synopsis
Unique challenges of fetal-brain MRI include successful acquisition of standard sagittal, coronal and axial views of the brain, as motion precludes acquisition of coherent orthogonal slice stacks. Technologists repeat scans numerous times by manually rotating slice prescriptions but inaccuracies in slice placement and intervening motion limit success. We propose a system to automatically prescribe slices based on the fetal-head pose as estimated by a neural network from a fast scout. The target sequence receives the head pose and acquires slices accordingly. We demonstrate automatic acquisition of standard anatomical views in-vivo.
Introduction
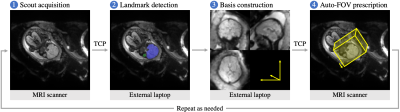
One of the unique challenges of fetal-brain MRI is the arbitrary orientation of the anatomy relative to the device axes, whereas clinical assessment requires standard sagittal, coronal and axial views relative to the fetal brain.1,2 Unfortunately, fetal motion limits acquisitions to thick slices which preclude retroactive resampling to provide standard planes (Figure 1). Throughout the session, technologists repeat acquisitions while incrementally adjusting the field of view (FOV), deducing the head pose from the previous stack until they obtain appropriately oriented images.Building on our prior work on landmark detection3 and automated prescription4, we present learning-driven FOV positioning and demonstrate automatic acquisition of standard anatomical planes of the fetal brain in-vivo.
Method
The system is fully integrated with a 3-T Siemens Skyra scanner, used to acquire all presented data with a flexible 60-channel body array. A laptop connected to the local sub-network is required. All communications are automatic.Setup
A deep neural network delineates the fetal brain and eyes in a full-uterus scout, from which the head pose is derived. A laptop receives the scout via TCP and returns coordinates to the target sequence (Figure 2). First, a scout is acquired and provided to the network for landmark detection. Second, the system resamples this image in the anatomical frame and displays it on the laptop, enabling the user to review anatomical alignment and repeat the scout if needed. Third, the user starts the target sequence, selecting sagittal, coronal or axial slices, which are positioned automatically.
Pulse sequences
The scout is a stack of 90 interleaved 3-mm GE-EPI slices with 128×128 matrix, (3 mm)2 resolution, no gap, TR/TA 6000 ms, TE 30 ms, FA 90°, 5/8 partial Fourier. The anatomical scan is a stack of 35 interleaved 3-mm HASTE5 slices with 256×256 matrix, (1.25 mm)2 resolution, no gap, TA 1:05 min, TR/TE 1800/100 ms, FA 160°, 5/8 partial Fourier, GRAPPA 2.
Landmark detection
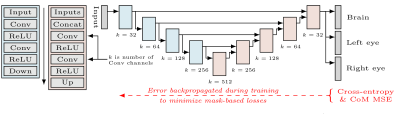
Brain and eye masks are estimated from input images using a U-Net6 (Figure 3) trained beforehand on a manually annotated dataset including 105 EPI stacks from 78 fetuses (parameters as above, except TR/TE 7000/36 ms). At each training step, images are augmented with random rotations and translations (0-10 voxels). We fit network parameters using a cross-entropy loss for each landmark and MSE for their barycenters. Brain-mask losses are weighted five times higher than losses for the eyes. Barycentric losses are scaled by 10-4 to maintain stability.
Anatomical frame
From the barycenters $$$\{c_b,c_l,c_r\}$$$ of the brain and eyes, we derive an orthonormal left-posterior-superior basis $$$F=\{u_l,u_p,u_s\}$$$.3 An initial right-left axis $$$\tilde{u}_l=(c_l-c_r)\,||c_l-c_r||^{-1}$$$ passes through the eyes. The mid-point $$$c_m=(c_l+c_r)/2$$$ and $$$c_b$$$ define an anterior-posterior axis $$$\tilde{u}_p=(c_b-c_m)\,||c_b-c_m||^{-1}$$$, and we let $$$\tilde{u}_s=\tilde{u}_l{\times}\tilde{u}_p$$$. As $$$\tilde{u}_l$$$ and $$$\tilde{u}_p$$$ may not be orthogonal, we use $$$u_l=\tilde{u}_p{\times}\tilde{u}_s$$$. The plane defined by $$$\{c_b,c_l,c_r\}$$$ is usually tilted from the axial plane by $$$\theta\approx-30^\circ$$$ about $$$\tilde{u}_l$$$: we choose as basis $$$F=R_l(\theta)\,(u_l\,\tilde{u}_p\,\tilde{u}_s)$$$, where $$$R_l$$$ is a rotation matrix about $$$u_l$$$.
Field of view
We prescribe slices in the anatomical frame by centering the FOV on $$$c_b$$$ and selecting the slice-normal vector from $$$\{u_l,u_p,u_s\}$$$ depending on the user's selection. The remaining vectors define the phase and read-out directions: to minimize potential wrapping4 we choose as vulnerable phase direction the vector with the smaller component along the maternal head-foot axis $$$z$$$.
Results
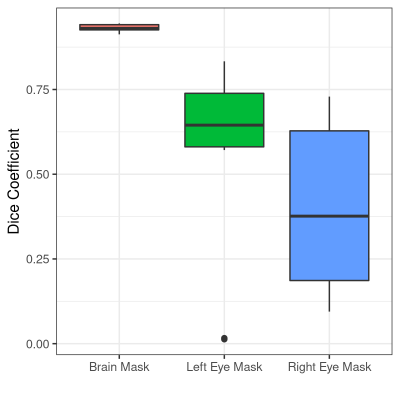
Landmark detectionWe test offline landmark detection on a held-out validation set of 10 EPI stacks that do not include the training subjects (Figure 4). Dice scores are (93.1±1.1)%, (56.1±30.1)%, and (41.0±25.0)% for the brain, left and right eye, respectively. The detection takes (4.8±0.6) sec on a laptop with a 2.8-GHz quad-core Intel i7-7700HQ CPU and 16 GB RAM. This reduces to (0.3±0.1) sec on an Nvidia V100-SXM2 GPU with 32 GB memory.
In-vivo evaluation
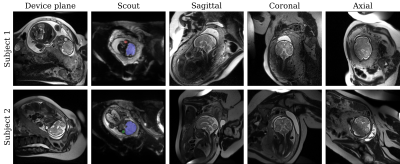
We perform automatic slice prescription in two healthy fetuses at 32 and 31 weeks' gestation (Figure 5). Landmarks are robustly identified for the first fetus. For the second, we repeat several scouts until the network detects the eyes, due to fetal motion and wrapping.
Discussion
Network robustnessAlthough the detection is less reliable in the second fetus, the real-time feedback enables the operator to repeat the scout until the head-pose is identified. We plan to improve the detection by increasing the training set and augmentation, e.g. with random image histogram scaling.
Scout acquisition
We will develop an optimized ~1-second scout with lower spatial resolution and a larger FOV, thus reducing between-slice motion and phase-wrap artifacts which impinge on the landmark detection.
Other applications
Auto-FOV positioning may prove useful in other applications of MRI, such as musculoskeletal MRI, where many structures are obliquely oriented to the standard planes. For example, capturing the thin articular cartilage of the hip and ankle can be challenging with 2D sequences,7 which remain the preferred clinical acquisition. Training networks for these body parts will extend our framework to aid technologists in prescribing difficult views.
Conclusion
We propose a framework for automated FOV and slice prescription of fetal-brain MRI and demonstrate its utility in-vivo. The modular design makes it extendable for users interested in other anatomy demanding oblique imaging.Acknowledgements
The authors thank Paul Wighton and Robert Frost for sharing code, and Andrew Hoopes for figure design inspiration. This work uses human subject data and is formally reviewed by the institution’s Human Research Committee. Support was provided in part by NIH grants NICHD K99 HD101553, U01 HD087211, R01 HD100009, R01HD093578 and NIBIB NAC P41 EB015902.References
1. Gholipour A et al. Fetal MRI: A technical update with educational aspirations. Concepts Magn. Reson. Part A. 2014;43(6):237–266.
2. Saleem SN et al. Fetal MRI: An approach to practice: A review. J. Adv. Res. 2014;5(5):507-523.
3. Hoffmann M et al. Improved, rapid fetal-brain localization and orientation detection for auto-slice prescription. ISMRM, Montreal, QC, Canada. 2019.
4. Hoffmann M et al. Fast, automated slice prescription of standard anatomical planes for fetal brain MRI. ISMRM, Paris, France. 2018.
5. Kiefer B et al. Image acquisition in a second with half-Fourier acquired single shot turbo spin echo. JMRI. 1994;4(86):86–87.
6. Ronneberger O et al. U-Net: Convolutional Networks for Biomedical Image Segmentation. CoRR. 2015;234-241.
7. Naraghi A and White LM. Three-dimensional MRI of the musculoskeletal system. AJR. 2012;199(3):W283-W293.
Figures