1326
Optimising spiral diffusion tensor cardiovascular magnetic resonance for high resolution ex-vivo STEAM imaging on a clinical scanner1National Heart and Lung Institute, Imperial College London, London, United Kingdom, 2Cardiovascular Magnetic Resonance Unit, Royal Brompton Hospital, London, United Kingdom
Synopsis
We demonstrate STEAM spiral diffusion tensor cardiovascular magnetic resonance (DT-CMR) for high resolution ex-vivo imaging (1x1x2mm2) on a clinical 3T scanner. We optimized the coil combination method, diffusion weighting, number of spiral interleaves and averages. A comparison in ex-vivo porcine myocardium shows substantial improvements in image quality when using the spiral method over a single-shot STEAM EPI protocol acquired with matched duration, spatial resolution and diffusion encoding.
Introduction
Ex-vivo Diffusion Tensor Cardiac Magnetic Resonance (DT-CMR) is a useful tool in investigating microstructure without confounding effects or in validating in vivo methods1. A number of studies have studied large mammalian hearts in clinical scanners, but resolution is often limited2 or sequences deviate from those available in vivo3. Recent studies demonstrated that segmented spiral STEAM provides high resolution in-vivo DT-CMR4. Here, we optimise interleaved STEAM spiral acquisitions for high resolution ex-vivo DT-CMR on a clinical scanner.Methods
Single slice mid-ventricular interleaved spiral STEAM DT-CMR was performed in a fixed porcine heart on a clinical 3T Siemens Vida. The heart was suspended in Fomblin and imaged in a head coil at 1x1x2mm2 resolution, 300ms diffusion time, TE=22ms,TR=1500ms, 6 diffusion directions, 20-40 interleaves/image, 4.9-9.5ms spiral duration, variable density spirals with linearly reducing field of view 360x360-180x180mm2. Coil sensitivity profiles, number of averages/interleaves and diffusion weighting (b-value) were optimised.Complex averaging was performed on matching b-values and diffusion encoding directions. DT-CMR data was processed using an in-house Matlab tool. Results are compared visually and via quality metrics: the standard deviation of the transverse angle over the ventricular myocardium (TA-std, increases with poorer quality); and R2 of the transmural change in HA (HA-R2, reduces with poorer quality)5,6
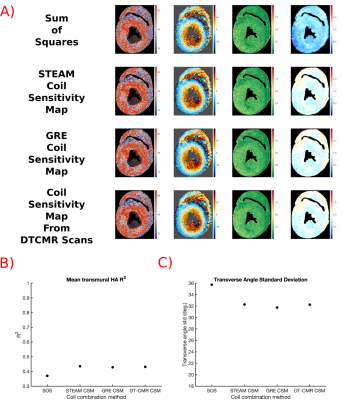
Optimal Coil Combination Method:
Coil sensitivity weighted coil combination results in superior SNR compared to Sum of Squares (SOS) but require the acquisition of Coil Sensitivity Maps (CSM). We compared 4 approaches (SOS, CSM acquired using: STEAM, gradient echo, and obtained directly from imaging data). 30xb=600smm-2 and 5xb=100smm-2 to identify the optimal strategy.
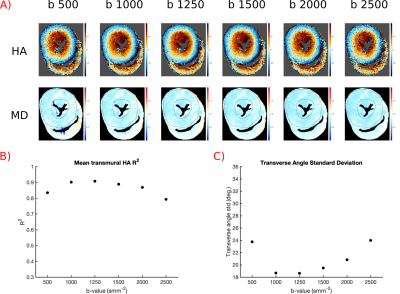
Optimise b-value:
Data was acquired with 30 averages of b=500,1000,1250,1500,2000,2500smm-2 and 5xb=100smm-2 to investigate the effect of b-value.
Optimise Averages/Interleaves:
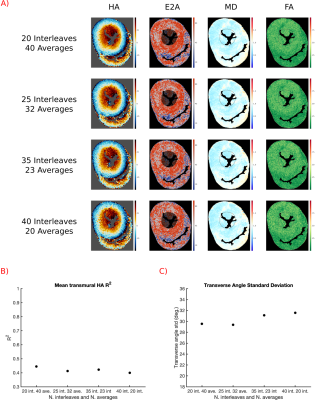
To optimise combination of averages and number of spiral interleaves, data was acquired with 50xb=1250smm-2 and 15xb=100smm-2 for spirals with 20-40 interleaves. For each datasets DT-CMR results were calculated with varying averages.
Compare the Optimal Spiral Protocol to EPI:
For comparison with a STEAM EPI method, the following optimised spiral protocol was used: 20 interleaves, 40xb=1250smm-2 and 7xb=100smm-2, CSM obtained directly from imaging data, 2hour duration. A matched duration single-shot STEAM EPI with 1x1x2mm3 resolution, 700xb=1250smm-2 and 120xb=100smm-2,6 diffusion directions, 300ms diffusion time, TE=112ms, TR=1500ms.
Results
Figure 1 shows results for the coil combination strategies. Maps appear similar and both HA-R2 and TA-std are similar for all techniques apart from SOS, suggesting that CSM can be obtained from the imaging data without penalties.Figure 2 shows results of varying b-value. HA appears least noisy and quality metrics are best in datasets with b=1000smm-2 and b=1250smm-2.
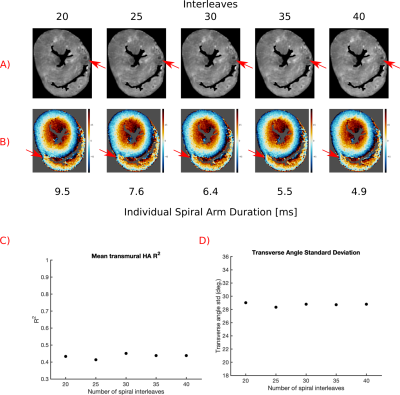
Figure 3 shows results of increasing the number of spiral interleaves. An off-resonance artefact caused by air or thrombus (Figure 3.A) decreases in size with increasing interleaves (shorter spiral interleaves), but there is little change in the associated HA or quality metrics.
Figure 4 shows the results of maintaining acquisition duration while varying the number of interleaves and averages. Visually, HA maps appear smoother with more averages (higher SNR) and a decreasing number of interleaves and quality metrics are improved.
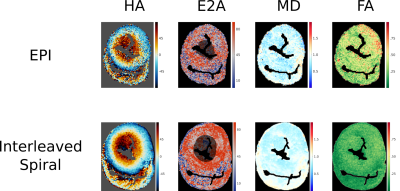
Figure 5 compares results from the EPI and spiral acquisitions for a matched 2hour scan. The results from the EPI appear noisy relative to the spiral acquisition.
Discussion
We have demonstrated that an interleaved spiral STEAM sequence is able to provide high resolution (1x1x2mm3) DT-CMR data with the long diffusion times frequently used for in-vivo acquisitions.The acquisition used to produce CSM has little effect on DT-CMR data quality. Extracting the CSM directly from the DT-CMR data, avoids additional acquisitions and the diffusion weighting does not appear to affect the coil combination’s performance.
Quality metrics were best and HA maps appear smoothest when b=1000-1250smm-2. Previous studies have suggested that b=1.11/MD is optimal7, which is b=1337smm-2 in this data (MD=0.83x10-3mm2s-1). At lower b-values the image noise is a substantial proportion of the signal loss and at higher b-values the diffusion weighted signal may approach the noise floor.
Longer spirals readouts result in more severe off resonance artefacts (Figure 3), but more data can be acquired in each TR, increasing SNR efficiency. Our results suggest that increasing the number of averages is preferential to increasing the number of spiral interleaves.
Based on these tests we selected an optimal protocol of b=1250 smm-2, CSM calculated from imaging data, 20 interleaves and 40 averages. Using this a single slice DT-CMR acquisition can be acquired within 2hours.
Spiral trajectories are well-suited to interleaved imaging and comparing our 2hour spiral acquisition to a time, b-value and resolution matched single-shot EPI sequence at high resolutions (1x1x2mm3) the spiral method yielded superior results. In future work, slice interleaving could enable equivalent DT-CMR images in ~25mins/slice.
Conclusion
Interleaved spiral trajectories can be used to acquire high quality ex-vivo DT-CMR at high resolution on a clinical scanner with the advantages of the long diffusion time of STEAM probing longer diffusion distances than spin-echo methods. Acquisitions are, however, time consuming and in this work, we investigated a number of optimisations to improve efficiency. Such methods will be valuable in assessing focal diseases, right ventricle, atria and thinned myocardium in chronic myocardial infarction.Acknowledgements
This work was funded by British Heart Foundation Grants:
RG/19/1/34160
FS/16/40/32167
FS/11/38/28864
References
1.Nielles-Vallespin, S. et al. Assessment of Myocardial Microstructural Dynamics by In Vivo Diffusion Tensor Cardiac Magnetic Resonance. J. Am. Coll. Cardiol. 69, 661–676 (2017).
2.Tous, C., Gentles, T. L., Young, A. A. & Pontré, B. P. Ex vivo cardiovascular magnetic resonance diffusion weighted imaging in congenital heart disease, an insight into the microstructures of tetralogy of Fallot, biventricular and univentricular systemic right ventricle. J. Cardiovasc. Magn. Reson. 22, 1–12 (2020).
3.Pashakhanloo, F. et al. Submillimeter diffusion tensor imaging and late gadolinium enhancement cardiovascular magnetic resonance of chronic myocardial infarction. J. Cardiovasc. Magn. Reson. 19, 1–14 (2017).
4.Gorodezky, M. et al. High resolution in-vivo DT-CMR using an interleaved variable density spiral STEAM sequence. Magn. Reson. Med. 81, 1580–1594 (2019).
5.Scott, A. D. et al. An in-vivo comparison of stimulated-echo and motion compensated spin-echo sequences for 3 T diffusion tensor cardiovascular magnetic resonance at multiple cardiac phases. J. Cardiovasc. Magn. Reson. 20, 1–15 (2018).
6.von Deuster, C., Stoeck, C. T., Genet, M., Atkinson, D. & Kozerke, S. Spin echo versus stimulated echo diffusion tensor imaging of the in vivo human heart. Magn. Reson. Med. 76, 862–872 (2016).
7.Jones, D. K. & Basser, P. J. ‘Squashing peanuts and smashing pumpkins’: How noise distorts diffusion-weighted MR data. Magn. Reson. Med. 52, 979–993 (2004).
Figures