1325
Highly Accelerated Multi-shot EPI based Diffusion MRI Using SMS and Joint k-q Under-sampling Enabled Using Deep Learned Manifold Priors1University of Iowa, Iowa City, IA, United States
Synopsis
We propose a new acceleration and reconstruction method for highly accelerated multi-shot dMRI. The acceleration makes use of multi-band excitation in combination with joint k-q undersampling. We develop new iterative reconstruction with deep learned q-space manifold priors to enable the recovery for the DWIs from 12-fold under-sampled data. The reconstruction error is shown to be less than 3%. The proposed method enables utilization of multi-shot EPI trajectories for diffusion microstructure and connectivity studies requiring high q-space coverage, without prolonging scan time.
Introduction
Diffusion MRI (dMRI) is a widely used neuroimaging technique that provides a non-invasive means to study the brain in detail including its connectivity, and microstructural changes in response to neurological disorders. While single-shot echo-planar (ssEPI) methods have been the main workhorse for dMRI, the low resolution of ssEPI is a major drawback of the technique. The poor resolution is a consequence of the long echo-time associated with the ssEPI readout, which limits its k-space coverage. Low resolution leads to partial volume confounds in both connectivity and microstructural studies.Multi-shot echo-planar (msEPI) methods can provide improved spatial resolution and can reduce the partial volume artifacts. Combining msEPI-based dMRI (msdMRI) with high q-space coverage can thus enable microstructural and connectivity studies with improved accuracy. However, such studies are only practical if high acceleration factors ( > number of shots) can be achieved. Previously, simultaneous multi-slice imaging (SMS) was employed to speed up the acquisition of msdMRI1-3. In this work, we propose to further speed up the acquisition of SMS-accelerated msdMRI using joint k-q under-sampling4. We extend our previous model-based reconstruction employing learned priors in q-space5 to accommodate SMS and joint k-q acceleration.
Methods
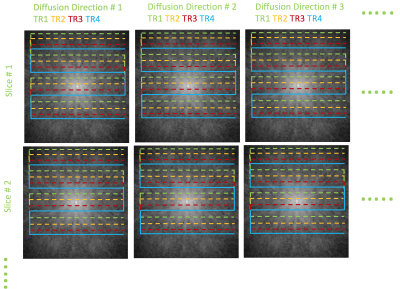
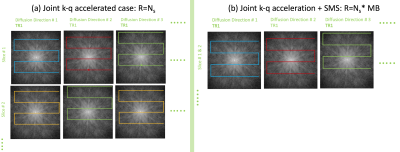
Figures 1-2 illustrates the proposed acceleration scheme combining SMS and joint k-q under-sampling. Specifically, we randomize the k-q space sampling of the combined data from multiple slices, using a highly accelerated msEPI trajectory, leading to an acceleration factor $$$MBxN_s$$$. However, the reconstruction of the above highly under-sampled data is challenging. Simple parallel imaging methods are inadequate to reconstruct the individual DWIs from the highly under-sampled data in addition to separating the aliased slices.We propose to augment the current parallel imaging methods with additional priors in q-space to aid the above recovery. Moreover, using a q-space prior, we devise a joint reconstruction of all the diffusion directions, simultaneously. The joint recovery becomes a powerful strategy to maximally exploit the redundancy present in the measured k-q samples. Such an approach will fully exploit the random under-sampling pattern which is meant to distribute the aliasing patterns of the DWIs to different spatial locations. Specifically, the joint reconstruction is given by:
$$\mathbf{S^{^*}}\!\!~=~argmin_{_{\tilde{\mathbf{S}}}}||\mathcal{A}({\mathbf{S}})-\widehat{\mathbf{Y}}||_2^2~+~\lambda_1~||\mathcal{P}_{\Theta}(\mathbf{S})||_2^2 +~\lambda_2~||\mathbf{S}||_{TV}~,~~~~~~~~~~~~[1]$$
where
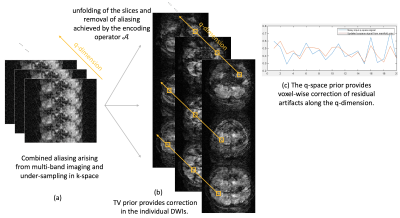
$$\mathcal{P}_{\Theta}(\mathbf{S})~=\mathcal{D}_{\Theta}(\mathbf S) -\mathbf{S}.$$
Here, $$$\mathbf{S}$$$ is the set of DWIs to be reconstructed from all the directions and the simultaneously excited slices, and $$$\widehat{\mathbf{Y}}$$$ is the multi-channel k-space measurements. The first term in Eq [1] enforces data consistency to the k-space measurements and performs slice unalisaing using the multi-channel encoding operator $$$\mathcal{A}$$$. The second term introduces the q-space prior. Specifically, we make use of a q-space manifold prior, where we pre-learn the signal manifold corresponding to the diffusion acquisition. We achieve this using a biophysical modeling approach, where appropriate biophysical models that are general enough to represent in-vivo diffusion signals, are used to define the q-space manifold. The manifold is learned using deep learning by training a denoising autoencoder neural network. Training data to learn the manifold are derived from the biophysical models and do not involve in-vivo training data. This biophysics-driven training framework not only circumvents the need to generate pristine in-vivo training data, it also allows modifications of acquisition patterns as needed. For example, the same trained network can be re-used for any acceleration factor MB x Ns, which is hard to achieve with data-driven training frameworks.
We denote the pre-learned q-space manifold as $$$\mathcal{D}_{\Theta}(\mathbf S)$$$ and incorporate this into a traditional iterative reconstruction. In Eq [1], we use the manifold prior to minimize the projection error $$$\mathcal{P}_{\Theta}(\mathbf{S})$$$ to the q-space manifold. This voxel-wise projection of the diffusion signal to the pre-learned q-space manifold enables the joint recovery of the DWIs for all the slices involved. The projection error is minimized subject to data-consistency to the measured data. Additional image domain prior is imposed using of total variation (TV). A schematic of the above recovery is shown in figure 3.
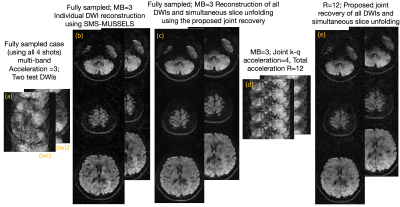
To test the proposed acceleration and reconstruction scheme, we performed an in-vivo dMRI experiment using a multi-band EPI sequence. Data was collected using a 32-channel coil on a 3T MRI with Gmax of 33mT/m. Using blipped-CAIPI technique, a MB=3 was achieved. The k-space sampling was performed using a 4-shot EPI sequence for b-value=1000s/mm2, 20 diffusion directions. This fully sampled data was retrospectively under-sampled in the k-q domain. For every diffusion direction, only 1 random k-space shot was retained, giving a total acceleration of 12 (MB=3,Ns=4).
Results
Figure 4 shows the recovery of DWIs from the proposed method for two test diffusion directions for the three slices that were simultaneously excited. For comparison, the reconstruction using the fully sampled data is also provided.Discussion
Notably, the reconstruction of the fully sampled case is also improved using the joint recovery method compared to the individual DWI reconstruction. The joint reconstruction method shows minimal artifacts at R=12 compared to the SMS only accelerated case (R=3). The RMSE is close to 2.6% confirming reasonable accuracy for the proposed method.Conclusion
The combined acceleration provided by SMS + joint k-q under-sampling offers the necessary speed up required to employ msEPI methods for diffusion microstructure and connectivity studies involving extensive q-space measurements. The joint recovery method is shown to provide reasonable reconstruction of such highly accelerated data.Acknowledgements
Financial support for this study was provided by grants NIH 5 R01 EB022019, 5 R01 MH111578, NIH 1R01EB019961-01A1. This work was conducted on MRI instruments funded by 1S10OD025025-01 and 1S10RR028821-01.References
1.Dai, E., Ma, X., Zhang, Z., Yuan, C. and Guo, H. (2017), Simultaneous multislice accelerated interleaved EPI DWI using generalized blipped‐CAIPI acquisition and 3D K‐space reconstruction. Magnetic Resonance in Medicine, 77: 1593-1605.
2. B. Bilgic et al., “Highly accelerated multishot echo planar imaging through synergistic machine learning and joint reconstruction,” Magnetic Resonance in Medicine., 2019; 82: 1343– 1358.
3. M. Mani et al., “SMS MUSSELS: A navigator-free reconstruction for simultaneous multi-slice-accelerated multi-shot diffusion weighted imaging,” Magnetic Resonance in Medicine, 2020; 83: 154– 169.
4. M. Mani, M. Jacob, A. Guidon, V. Magnotta, and J. Zhong, “Acceleration of high angular and spatial resolution diffusion imaging using compressed sensing with multichannel spiral data,” Magnetic Resonance in Medicine, 2015; 73: 126–138.
5. Mani MP, Aggarwal HK, Ghosh S, and Jacob M. Model-Based Deep Learning for Reconstruction of Joint k-q Under-sampled High Resolution Diffusion MRI. In IEEE International Symposium on Biomedical Imaging (ISBI), 2020.
Figures