1321
Simple improvement of Multi-Dimensional diffusion MRI(MD-dMRI) image quality by double-sampled EPI
Nicolas Geades1, Oscar Jalnefjord2,3, Guillaume Gilbert4, and Maria Ljungberg2,3
1MR Clinical Science, Philips, Gothenburg, Sweden, 2Department of Medical Physics and Biomedical Engineering, Sahlgrenska University Hospital, Gothenburg, Sweden, 3Department of Radiation Physics, University of Gothenburg, Gothenburg, Sweden, 4MR Clinical Science, Philips, Markham, ON, Canada
1MR Clinical Science, Philips, Gothenburg, Sweden, 2Department of Medical Physics and Biomedical Engineering, Sahlgrenska University Hospital, Gothenburg, Sweden, 3Department of Radiation Physics, University of Gothenburg, Gothenburg, Sweden, 4MR Clinical Science, Philips, Markham, ON, Canada
Synopsis
This study presents a novel implementation of established methods to eliminate ghosting artifacts in Multi Dimensional diffusion MRI (MD-dMRI) images. By replacing scan time used for acquiring images purely used for averaging (when applicable), with EPI acquisitions at opposing readout directions, ghost artifacts can be eliminated. The implementation provides ghost-free images with a small increase in SNR and a minimal (7%) increase in scan time.
Introduction
Recently Multi-Dimensional Diffusion Imaging (MD-dMRI) has transformed the field of diffusion MRI by offering non-invasive characterization of complex anisotropic and heterogeneous structures like the human brain in extraordinary detail [1]. The very nature of MD-dMRI requires lengthy scan times and collection of a large amount of diffusion-weighted images that provide enough information for a good model fit [2]. In order to achieve this large amount of data collection in a reasonable amount of time, multiple acceleration methods are used like parallel imaging (SENSE), multi-band/multi-slice and an EPI readout, but these acceleration methods come with associated artifacts like geometric distortions and nyquist ghosting (figure 1). This work presents how the long established method of acquiring the EPI data at both, opposing readout directions [3], together with the nature of multiple averages required for MD-dMRI data, can result in ghost-free images with potentially no increase in scan time. While this ghost correction method [3] is not new, its use has been rather limited due to the added scan time. Our proposed implementation is novel and solves a very common problem with MD-dMRI, especially when used for brain tumor imaging where the artifacts can obscure the tumor tissue.Methods
Data acquisition and subjects: All scanning was performed on a Philips Achieva dS 3T system in a healthy volunteer with ethical approval. The protocol covers the brain (240mm x 240mm x 120mm), at 2.5mm isotropic resolution, with SENSE factor 1.9 and Multi-Band (MB) SENSE factor 2, with an EPI readout. The TR and TE are highly dependent on the diffusion encoding gradients and MB factor used, and in this case were set to TR = 5700ms and TE = 108ms. The protocol includes two separate scans, one for spherical diffusion encoding and one for linear diffusion encoding (figure 2) with a scan time of 4minutes and 4minutes 32seconds respectively. In our protocol, b-values of 100, 700, 1400 and 2000 s/mm2 are used, with multiple directions/averages acquired for each b-value.Nyquist ghost elimination at low time penalty: The MD-dMRI protocol acquires images at various b-values and multiple averages, in order to reduce noise and fit residuals, but the b-value averages can be replaced by double measurements at opposing readout directions [3] (in this work denoted EPI Both Signs(EBS)). In the case of spherical encoding, images have no directional resolution (neglecting higher order frequency effects) and hence replacing averages with EBS results in no additional added scan time. In the case of linear encoding, at least 6 directions are required to produce isotropic diffusion images but more images are often acquired for noise reduction, especially at higher b values. In the work shown in this abstract, we only acquire six images/directions for the b = 100 s/mm2 data so using EBS to eliminate ghosting in those images results in 6 more acquired images at a TR of 5700ms, and a subsequent 34s increase in scan time. The total scan time with and without EBS is 9 minutes 6seconds and 8 minutes 32 seconds respectively (<7% increase with EBS ON).
The strength of the ghosting artifact is reduced with increasing b-value (figure 3). This suggests that the artifact source is the cerebrospinal fluid (CSF) signal that is not suppressed in low b-value images. The EBS method is usually not used for Diffusion Weighted Imaging (DWI) as it doubles the acquisition time, except for the b=0 image where both SNR and directional resolution are not an issue. In this work we enable the option to use EBS on user-selected b-value images.
Results
The feasibility of this method was successfully demonstrated on a healthy volunteer, acquiring ghost-free trace images and MD-dMRI maps by enabling EBS on only the b=100s/mm2 images. Figure 1 shows the extent of the artifacts under regular circumstances. Figure 3 shows the effects of EBS on b = 100, b = 400, b = 700 and b = 1000 s/mm2 images. Figure 4 shows MD-dMRI maps with EBS ON for b = 100 s/mm2 images only, versus MD-dMRI maps with EBS OFF.Discussion
A novel study is presented that uses an established methodology to improve the quality of MD-dMRI maps with minimal increase in scan time. The work shows that the main source of signal in the nyquist ghost is CSF, and by applying EBS only on diffusion-weighted images that show strong CSF signal, it is possible to remove the artifacts from the final images with a minimal increase in scan time. This work enables MD-dMRI images of great diagnostic quality, especially when combined with geometric distortion correction. The simple modification presented here eases some of the strict requirements of MD-dMRI on system hardware and increases confidence in the results.conclusions
This work shows that it is possible to get artifact-free diffusion weighted images for MD-dMRI at the same or higher SNR, with a minimal (7%) increase in scan time. This greatly improves the confidence of MD-dMRI results, especially when artifacts can obscure vital parts of tumors.Acknowledgements
This study was funded by grants from the Swedish Cancer Society (CAN 2018/628), the King Gustav V Jubilee Clinic Cancer Research Foundation (2018:217) and the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement (ALFGBG-825191)References
- D. Topgaard, Multidimensional diffusion MRI, Lund: Journal of Magnetic Resonnce, 2017.
- F. Szczepankiewicz, The link between diffusion MRI and tumor heterogeneity: Mapping cell, Lund: NeuroImage, 2016.
- Q. X. e. a. Yang, Double-sampled echo-planar imaging at 3 tesla, Journal of Magnetic Resonance, 1996.
Figures
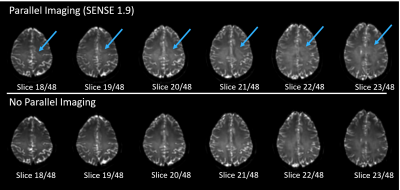
Ghost
artifacts visible when parallel imaging is used. b = 100 s/mm2
images shown
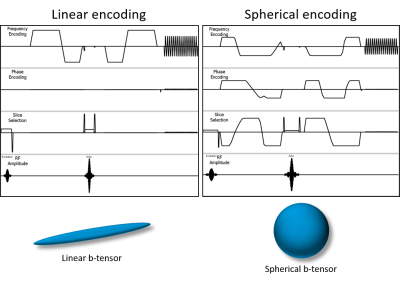
Linear
and Spherical diffusion encoding diagrams (not to scale), showing the diffusion
encoding gradients, EPI readout and corresponding linear and spherical
b-tensors
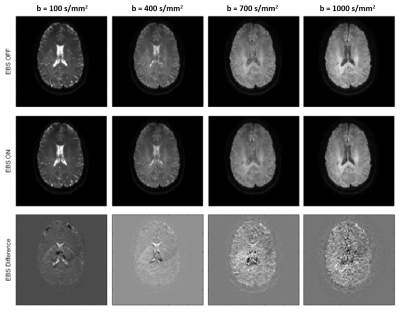
Artifact
vs. b-value. Images are shown with EBS on (top row) and visible ghosting
artifacts, EBS off (middle row) with no visible artifacts, as well as a
difference image (bottom row)
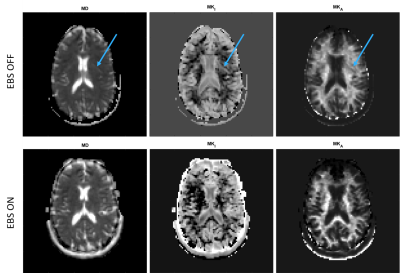
MD-dMRI maps with and without EBS. Mean Diffusivity (MD),
Mean Isotropic Kurtosis (MKI), Mean Anisotropic Kurtosis (MKA)