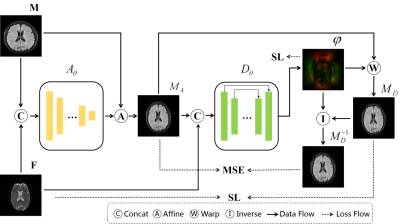1250
Stroke analysis with fully automatic multi-contrast MR image registration1Paul C Lauterbur Research Center, Shenzhen Institutes of Advanced Technology Chinese Academy of Sciences, Shenzhen, China, 2Radiology department, Peking University Shenzhen Hospital, shenzhen, China, 3United Imaging Research Institute of Innovative Medical Equipment, Shenzhen, China, 4Pengcheng Laboratory, Shenzhen, China
Synopsis
Stroke is a leading cause of death and disability worldwide. However, the misalignment between multi-contrast MR images bring difficulties in identifying the lesions. We propose an automatic framework including affine and deformation transformation for multi-contrast stroke images registration. In the framework, a new inverse operation is proposed to maintain the topology of images and a background suppression loss function is designed to optimize background predictions. The method achieves the best Dice score of 0.826 compared to 5 state-of-the-art methods. Moreover, our method is about 17 times faster than the most competitive method SyN when testing on a same CPU.
Introduction
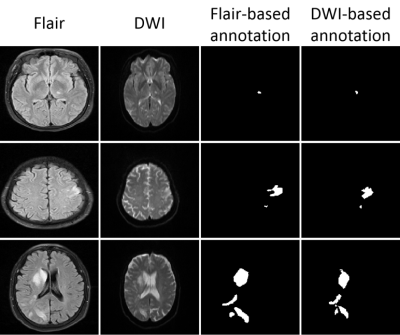
Stroke is the most common cerebrovascular disease [1]. Multi-contrast magnetic resonance imaging can help to detect strokes. However, due to 1) the different multi-contrast imaging parameters settings, 2) inevitable physiological activities and 3) different space coordinates for scanning [2-5], multi-contrast images may have large discrepancies(e.g. Fig. 1)causing difficulties in identifying stroke lesion quickly and accurately. Registration can be useful in handling the misalignment between different contrast images and therefore help identifying stroke lesions more efficiently [6-8]. Nevertheless, traditional multi-contrast registration algorithms commonly rely on an interactive optimization process, which is not suitable for time-sensitive diagnosis in clinical practice and bringing additional operations to computing large-scale analysis. Deep learning-based methods can speed up the registration process [9-11]. This study proposes a fully automatic multi-contrast MR image registration method for quickly identifying lesions between different contrasts.Method
The proposed method proposes multi-contrast stroke image registration. As shown in Figure 2. The proposed method consists of affine transformation network Aθ to solve the problem of different multi-contrast imaging parameters and deformable network Dθ to relieve the spatial inconsistency. Subsequently, a novel inverse transformation was proposed. This method calculates the inverse deformation field φ-1 from the deformation field φ by self-deformation and taking the opposite direction in tensor. We warp the φ-1 to moving image deformation MD and constrain it to be consistent with the input image MA through mean square error (MSE). Besides, a loss function (Suppress Loss, SL) is designed to suppress the misalignment of the background region. The formula is as follows:
$$
SL= \sum_{i}\begin {cases}αMI&(M_ {A_ {i}},M_ {D_ {i}}^{-1})+βSim(M_ {A_ {i}},M_{D_{i}}^{-1}),&\text{if $M_{A_{i}} $ <γ}\\&αMI(M_ {A_ {i}},M_ {D_ {i}}^{-1}), &\text{otherwise}\end {cases}
$$
Here, MI represents mutual information (MI) loss [12] and Sim represents the similarity function (we adopt MSE) in subsequent experiments. γ is the background threshold based on the data.
Results and discission
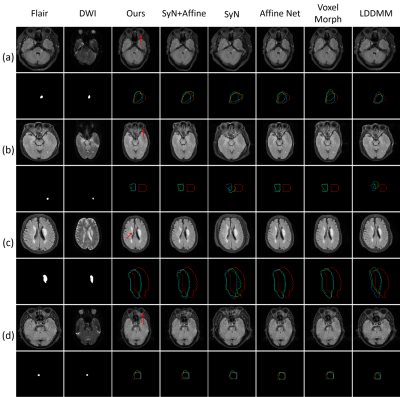
A total of 555 MR scanning cases were collected. Informed consent were obtained from each patient and the data were used under the constraint given by our ethics review board policy. Each case contains FLAIR and DWI contrast. Example predictions are shown in Fig. 3. Our method is not only more accurate in the registration of stroke lesions, other structures (red arrows), such as eyes and cerebrospinal fluid, but also show more reliable predictions.Our method achieves the highest stroke coincidence with Dice score of 0.826 as shown in Table 1. In addition, our method is approximate 17 times faster than the second-ranked SyN+Affine algorithm when testing on an ‘Intel(R) Xeon(R) Silver 4114’ CPU.
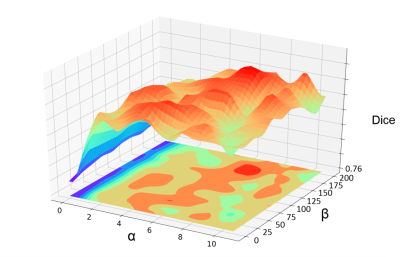
In order to verify the robustness of the proposed SL, we conducted a parameter comparison mentioned in eq.1. The visualization result is shown in Fig.4. Except for some extreme settings (α<2), the registration performance is quite robust with changing α and β values.
Conclusion
The study of stroke is crucial to the treatment and prognosis in the clinic. The misalignment between contrast images is the main obstacle in this process. This paper proposes a new registration framework to accurately and quickly register stroke Flair-DWI images. Experiments have proved that the proposed method takes account of accuracy and time consumption, and achieves the best registration performance. It is hoped to not only reduce the difficulty for clinicians to identify lesions, but also provide a robust standardized process for large-scale imaging analysis.Acknowledgements
This research was partly supported by Scientific and Technical Innovation 2030-"New Generation Artificial Intelligence" Project (2020AAA0104100, 2020AAA0104105), the National Natural Science Foundation of China (61871371, 81830056), Key-Area Research and Development Program of GuangDong Province (2018B010109009), the Basic Research Program of Shenzhen (JCYJ20180507182400762), Youth Innovation Promotion Association Program of Chinese Academy of Sciences (2019351).References
[1] Z. Liu, C. Cao, S. Ding, Z. Liu, T. Han, and S. Liu, “Towards clinical diagnosis: Automated stroke lesion segmentation on multi-spectral mr image using convolutional neural network,” IEEE Access, vol. 6, pp. 57 006–57 016, 2018.
[2] R. Zhang, L. Zhao, W. Lou, J. M. Abrigo, V. C. Mok, W. C. Chu, D. Wang, and L. Shi, “Automatic segmentation of acute ischemic stroke from dwi using 3-d fully convolutional densenets,” IEEE transactions on medical imaging, vol. 37, no. 9, pp. 2149–2160, 2018.
[3] D. L. Hill, P. G. Batchelor, M. Holden, and D. J. Hawkes, “Medical image registration,” Physics in medicine & biology, vol. 46, no. 3, p. R1, 2001.
[4] L. G. Brown, “A survey of image registration techniques,” ACM computing surveys (CSUR), vol. 24, no. 4, pp. 325–376, 1992.
[5] K. Van Leemput, F. Maes, D. Vandermeulen, and P. Suetens, “Automated model-based bias field correction of mr images of the brain,” IEEE transactions on medical imaging, vol. 18, no. 10, pp. 885–896, 1999.
[6] B. M. Dawant, “Non-rigid registration of medical images: purpose and methods, a short survey,” in Proceedings IEEE International Symposium on Biomedical Imaging. IEEE, 2002, pp. 465–468.
[7] E. Burke Quinlan, L. Dodakian, J. See, A. McKenzie, V. Le, M. Wojnowicz, B. Shahbaba, and S. C. Cramer, “Neural function, injury, and stroke subtype predict treatment gains after stroke,” Annals of neurology, vol. 77, no. 1, pp. 132–145, 2015.
[8] S.-L. Liew, J. M. Anglin, N. W. Banks, M. Sondag, K. L. Ito, H. Kim, J. Chan, J. Ito, C. Jung, N. Khoshab et al., “A large, open source dataset of stroke anatomical brain images and manual lesion segmentations,” Scientific data, vol. 5, p. 180011, 2018.
[9] M. Jaderberg, K. Simonyan, A. Zisserman et al., “Spatial transformer networks,” in Advances in neural information processing systems, 2015, pp. 2017–2025.
[10] G. Balakrishnan, A. Zhao, M. R. Sabuncu, J. Guttag, and A. V. Dalca, “An unsupervised learning model for deformable medical image registration,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 9252–9260.
[11] J. Krebs, H. Delingette, B. Mailhe, N. Ayache, and T. Mansi, “Learning a probabilistic model for diffeomorphic registration,” IEEE transactions on medical imaging, vol. 38, no. 9, pp. 2165–2176, 2019.
[12] F. Maes, A. Collignon, D. Vandermeulen, G. Marchal, and P. Suetens, “Multimodality image registration by maximization of mutual information,” IEEE transactions on Medical Imaging, vol. 16, no. 2, pp. 187– 198, 1997.
Figures