1243
Adaptive Multi-contrast MR Image Denoising based on a Residual U-Net using Noise Level Map1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
Synopsis
Multi-contrast MRI offers us images with complementary diagnostic information. Despite the dramatic difference in contrast, multi-contrast images often share highly correlated structure information. A deep learning (DL) based strategy is proposed to denoise multi-contrast MR images with flexible noise-levels using residual U-Net. This method utilizes the structural similarities across contrasts by simultaneously denoising multiple contrasts while existing single-contrast MRI denoising methods neglect the analogous structure information. The proposed method outperforms BM3D in terms of better noise reduction and details preservation. More importantly, we introduce a noise-level map that can be manually set to fit the different noise levels.
Introduction
Multi-contrast MRI is a useful technique to aid clinical diagnosis. It offers us multi-contrast images with complementary diagnostic information. Although the signal intensity varies dramatically across different contrasts, the underlying tissue often shares strong structural similarities or correlations. Most existing MRI denoising methods only carry out denoising on single contrast without using analogous structure information to support the restoration. Moreover, conventional deep learning (DL) based methods train a model for blindly denoising images with different noise levels, which will compromise the performance. Alternatively, the trained model is for a specific level of noise reduction, which means multiple models are needed for different noise levels. A single model that can be adjusted to fit images with varying noise levels can offer better performance and higher flexibility. This study proposes an adaptive DL-based strategy to simultaneously denoise multi-contrast MR images with different noise levels using only a single model, which is implemented by combining U-Net1 with ResNet2.Methods
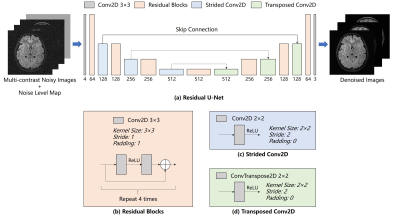
Proposed ModelMRI denoising recovers high-quality MR images y from the noisy MR images x. Generally, the neural network seeks a mapping function f that minimizes the difference between the denoised images and target noise-free images. The proposed method uses a residual U-Net architecture, which combines 4-scale U-Net and ResNet. ReLU activations are used after strided/transposed convolutional layers and between two convolutional layers within each residual block. All the convolutional layers are bias-free to impose scaling invariance.
Images of different contrasts are input as different channels. Inspired by FFDNet3 and DRUNet4, a noise level map is introduced as an additional input channel to balance noise reduction and details preservation. The noise level map can be manually adjusted to fit the input noise level, which is considered to be uniform within the FOV in this study. The model outputs denoised images as different channels.
Model Training and Evaluation
The network parameters were adjusted by minimizing the L1 loss between the denoised images and their ground-truths with Adam optimizer. A pre-trained model4 was used for initialization. 5800 multi-contrast image sets were selected from the HCP dataset. T1-weighted (T1w) images were acquired with MPRAGE with TR/TE/TI=2400/2.1/1000ms, flip angle (FA)=8°. T2-weighted (T2w) were acquired using 3D FSE with TR/TE=3200/565ms. All images have an isotropic resolution of 0.7×0.7×0.7mm3. T1w image, T2w image, and the averaged of T1w/T2w images were treated as three different contrasts. Noisy images were generated by adding complex white Gaussian noise with standard deviation (σ) ranging from 0 to 35 and used to train the proposed model.
The proposed method was also evaluated with human brain datasets acquired on a 3T Philips MRI scanner using a single-channel head coil. T1w images were acquired using 3D gradient-echo (GRE) with TR/TE=19/4ms, FA=30°, T2w images were acquired using 3D fast spin echo (FSE) with TR/TE=2500/213ms. T2-weighted FLAIR images were acquired with 3D FSE with TR/TI/TE=4800/1650/282ms. All images have an isotropic resolution of 1×1×1mm3. Complex white Gaussian noise of different levels was added to the reconstructed complex images. After that, the magnitude images were used for evaluating the proposed method.
Results
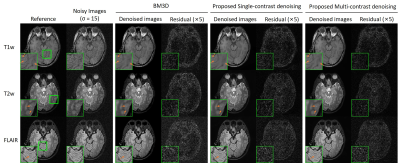
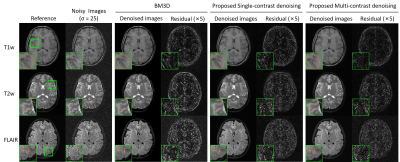
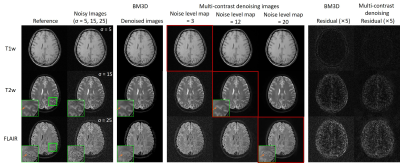
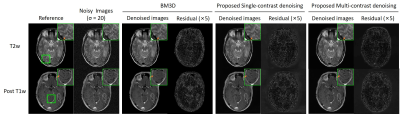
In Fig. 2, the same level of noise was added to the three images with different contrasts. Our approach preserved the structure details with noise significantly suppressed. Fig. 3 shows the robustness of the proposed method when images were corrupted with a much higher noise level. Fig. 4 indicates that our methods still performed well in noise reduction and details preservation when images of different contrasts have different noise levels. In this situation, we can adjust the noise level map to adapt the noise level of each contrast, so to achieve the best performance for all contrasts. Fig. 5 shows that even with pathology that manifested insufficient structural similarities across contrasts, the proposed method could remove noise with structural details well preserved. In contrast, BM3D removed the same level of noise but over-smoothed the image structures.Discussion and Conclusion
Our proposed denoising method utilizes the structural similarities by simultaneously denoising multiple contrasts using a residual U-Net. It shows a satisfactory performance in both noise reduction and details preservation at different noise levels. This is achieved because the noise level map can be manually adjusted to fit different noise levels. It's worth mentioning that in the presence of slight geometrical mismatches across different contrasts, the proposed method will still work because the receptive field of the model is large enough so that our extracted information can tolerate subtle geometrical mismatches. Note that images obtained with parallel imaging will have a spatially variant noise distribution. More importantly, such spatial variation can differ across different contrasts, as they may have non-identical sampling patterns. In the future, the model can be designed to have individual noise maps for each contrast to compensate for this issue.Acknowledgements
This study was supported by Hong Kong Research Grant Council (R7003-19, C7048-16G, HKU17112120, HKU17103819 and HKU17104020), Guangdong Key Technologies for Treatment of Brain Disorders (2018B030332001), and Guangdong Key Technologies for Alzheimer's Disease Diagnosis and Treatment (2018B030336001).References
1. Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
2. Kaiming He, Xiangyu Zhang, Shaoqing Ren, et al. "Deep residual learning for image recognition." Proceedings of the IEEE conference on computer vision and pattern recognition. 2016.
3. Kai Zhang, Wangmeng Zuo, Lei Zhang, et al. "FFDNet: Toward a fast and flexible solution for CNN-based image denoising." IEEE Transactions on Image Processing 27.9 (2018): 4608-4622.
4. Kai Zhang, Yawei Li, Wangmeng Zuo, et al. "Plug-and-play image restoration with deep denoiser prior." arXiv preprint arXiv:2008.13751 (2020).
5. Kostadin Dabov, Alessandro Foi, Vladimir Katkovnik, et al. "Image denoising by sparse 3-D transform-domain collaborative filtering." IEEE Transactions on image processing 16.8 (2007): 2080-2095.
Figures