1242
Multi-Contrast Whole Brain MRI: Optimization of Imaging Parameters and Motion Compensation1Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States
Synopsis
Multi-contrast anatomical brain MR images are widely used for tissue characterization and lesion detection, by applying multiple sequences. We developed a single continuous data acquisition applied with multiple inversion pulses, allowing multi-contrast imaging with high data acquisition efficiency. In order to achieve accurate tissue segmentation based on the multi-contrast images, imaging parameters are optimized based on simulated signal evolutions for achieving good image quality and accurate tissue characterization. A motion compensation approach for 3D whole-brain, multi-contrast MRI, was developed by exploiting the flexible data sampling strategy that allows for arbitrary combination of data collected throughout acquisition time and inversion times.
INTRODUCTION
Multi-contrast anatomical brain MR images are routinely acquired with multiple sequences for tissue characterization and lesion detection. Although a single continuous data acquisition applied with multiple inversion pulses allows for high data acquisition efficiency with multi-contrast imaging [1-3], imaging parameters need to be optimized and motion compensation applied in order to achieve an accurate tissue segmentation based on the multi-contrast images. This study investigates: 1) the simulated signal evolutions for optimizing imaging parameters for achieving good image quality and accurate tissue characterization, and 2) a motion compensation approach for 3D whole-brain, multi-contrast MRI, that exploits our flexible data sampling strategy and allows for the arbitrary combination of data collected throughout multiple inversion times.METHODS
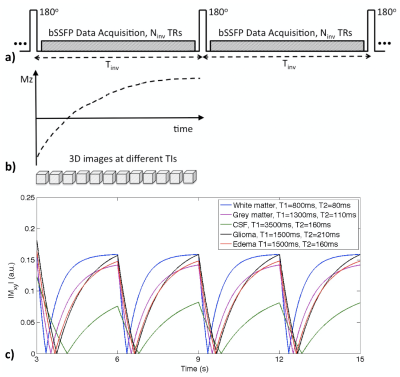
Subjects & Data Acquisition: Data were acquired on 6 patients with enhancing gliomas and 2 healthy volunteers, on a 3T MRI scanner (GE Healthcare, Milwaukee) with 32-channel head coil. Scan parameters were: FOV=25.6x19.2cm, image matrix = 256x192x160, FA=30o, TR/TE=4.2/1.7ms, BW=125kHz, Tinv=3s, and scan time of 6 mins. Multi-contrast images of isotropic 1mm resolution at 20 TIs were reconstructed using k-t SPARSE-SENSE method [5,6] (achieving a ~10-fold acceleration factor).Imaging Parameter Optimization: We developed single-scan multi-contrast whole brain MRI as demonstrated in Figure 1a, through applying a series of nonselective inversion pulses followed by a fixed number of segmented bSSFP acquisitions using an undersampling strategy designed for high acceleration [4] during a specific inversion recovery interval of Tinv. With a continuous data acquisition, a series of 3D images can be reconstructed at different inversion times (TIs) (Figure 1b). Simulated signal evolutions of different brain tissues with assumed T1/T2 values and given scan parameters are shown in Figure 1c. Imaging parameters were optimized to yield both good image quality and accurate segmenations of brain tissue.
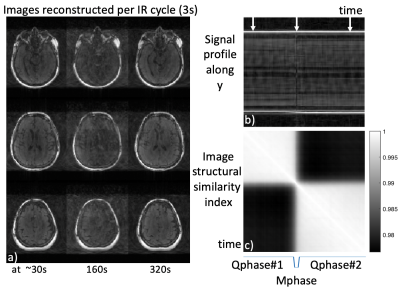
Motion Compensation: Although the variable density sampling strategy is inherently robust to small amounts of motion due to repeated acquisition of low frequency k-space, any excess motion may cause obvious degradation of image quality, especially for the case when data from each image volume (at any TI) is acquired during a series of inversion recovery cycles. We propose to retrospectively track the motion and performed motion compensation in the k-space domain before sorting k-space data for multi-contrast image reconstruction. To track the amount of motion, images with lower spatial resolution (2 mm isotropic) per Tinv were reconstructed throughout the 6-minute data acquisition window using a viewsharing method. The image structural similarity index (ISSI) was calculated among each 3D image volume with the other time points. ISSI was then utilized with k-means clustering to identify the quiescent periods (two were identified in current study) with minimal motion (data for motion correction and image reconstruction) as well as the motion-corrupted phase (data rejection, not included for reconstruction). The rigid transformation (translation and rotation) was calculated between two quiescent phases, and then applied to k-space data of the second quiescent phase for registration.
RESULTS
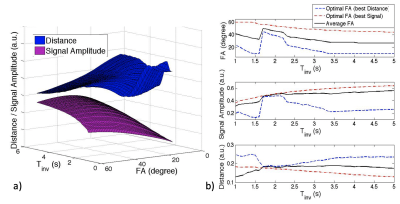
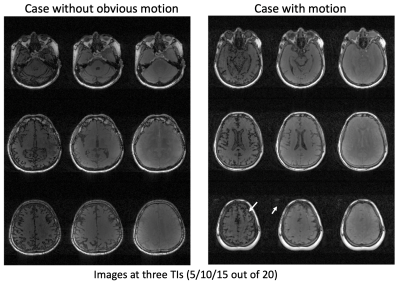
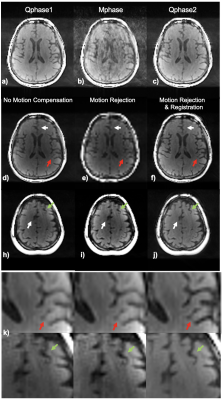
Image Parameter Optimization: Given a range of flip angles (10-60o) and different Tinv (1-5s), the averaged signal amplitudes of brain tissues (referring to Figure1c) are plotted as a 3D visualization (purple surface), as long as the averaged Euclidean distance between the signal evolutions of different tissues (blue surface) in Figure 2a. As seen in Figure 2b, there is a tradeoff for maximizing signal amplitude (corresponding to signal-to-noise level) versus distance (corresponding to tissue contrast). The average flip angles (black curve in Figure 2b) obtained based on maximizing signal (red curve) and distance (blue curve), respectively, are reasonable choices for joint optimization, resulting in an optimal FA of around 30o for a Tinv of 2.5-4s (Tinv=3s was chosen).Motion Compensation: Motion artifacts were observed in two of the patient scans. Figure 3 shows the images on three orthogonal planes (rows) from two patient scans (with and without obvious motion artifacts). Artifacts in brain tissue as well as in the background (arrows) are present when motion corruption happens (right block). The data of the case shown on the right of Figure 3 was used to generate a series of images, each per IR cycle (3s), as shown in Figure 4a (images at three representative time points are displayed). Motion occurred around 160s during the 6 minutes scan, as shown in Figure 4b (second arrow). The ISSI was calculated for each image volume with the other time points (shown in Figure 4c). Two quiescent phases and the motion corrupted phase were detected by applying k-means clustering on the ISSI. Figure 5 shows images of the detected three phases, without and with motion compensation. The images of Qphase2 were registered to the images of Qphase1 in kspace. The images with motion rejection and registration (data from Qphases1& registered Qphase2, Figure 5f&j) clearly show the improved sharpness (arrows) compared to the images without motion correction (Figure 5d&h) and with motion rejection only (Figure 5e&i). Enlarged regions illustrating these effects are also shown in Figure 5k).
CONCLUSIONS
We optimized the imaging parameters and implemented motion compensation for multi-contrast whole brain MRI which resulted in better quality images. Current work is underway to assess the benefit of these approaches on the accuracy of the resulting tissue segmentation.Acknowledgements
Research reported in this abstract was supported by NIH/NCIR21CA238137.References
1. Liu J, et al. 3D single-scan multi-contrast brain MRI for lesion detection in patients with gliomas without contrast administration. ISMRM Workshop on MR Value, March 2019.
2. Liu J, et al. 3D single-scan for multi-contrast-weighted MRI for lesion detection in patients with gliomas. SNO Annual Meeting 21(2019):NIMG- 45.
3. Cao P. et al. Simultaneous segmentation and relaxometry for MRI through multitask learning, Med Phys. 2019 Oct;46(10):4610-4621.
4. Liu J, et al. Accelerated MRI with CIRcular Cartesian UnderSampling (CIRCUS): a variable density Cartesian sampling strategy for compressed sensing and parallel imaging. Quant Imaging Med Surg 2014;4:57–67.
5. Otazo R, et al. Combination of compressed sensing and parallel imaging for highly accelerated first-pass cardiac perfusion MRI. Magn Reson Med. 2010 Sep;64(3):767-76.
6. Feng L, et al. MRM 2013. Highly accelerated real-time cardiac cine MRI using k-t SPARSE-SENSE. Magn Reson Med. 2013 Jul;70(1):64-74.
Figures